Locus Rank
78Gene
Id
230Gene Name
GPM6ADuplicated
TrueMaternal
FalseGene Not Within Locus (Nearby
FalseGene Description
Mouse Phenotype
Additional Image
Allen Brain

Allen Regions
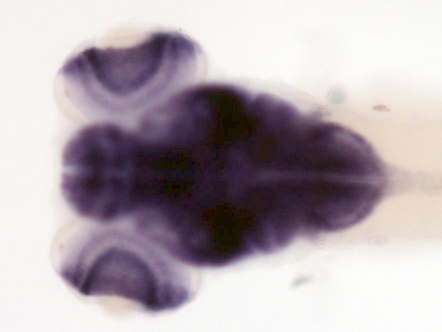
Zfin In Situ
Zfin Brain Areas
anterior lateral line ganglion, central nervous system, cranial ganglion, diencephalon, epiphysis, hindbrain, nucleus of the tract of the postoptic commissure, optic tectum, optic tract, optic vesicle, periventricular grey zone, posterior lateral line ganglion, retina, retinal ganglion cell layer, retinal inner nuclear layer, Rohon-Beard neuron, spinal cord, statoacoustic (VIII) ganglion, telencephalon, trigeminal ganglion, trigeminal placodePublished Zebrafish Pehnotype
Not In Allen Brain
FalseZFIN Link
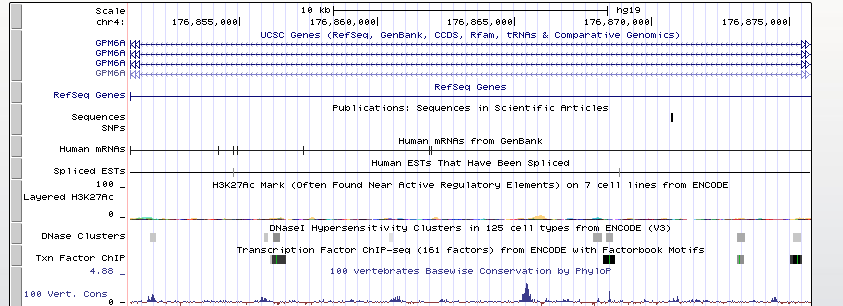
http://zfin.org/search?q=gpm6a&fq=category%3A%22Gene+%2F+Transcript%22&category=Gene+%2F+Transcript&fq=type%3A%22Gene%22Allen Link
http://mouse.brain-map.org/gene/show/87807More Additional Images
Papers
http://www.ncbi.nlm.nih.gov/pubmed/25242528Locus Rank
Locus Rank
78Genes in Region
GPM6AAssociated Snps
rs1106568GWAS Region (hg19)
Genes Skipping
Ricopili Plot Surrounding Area
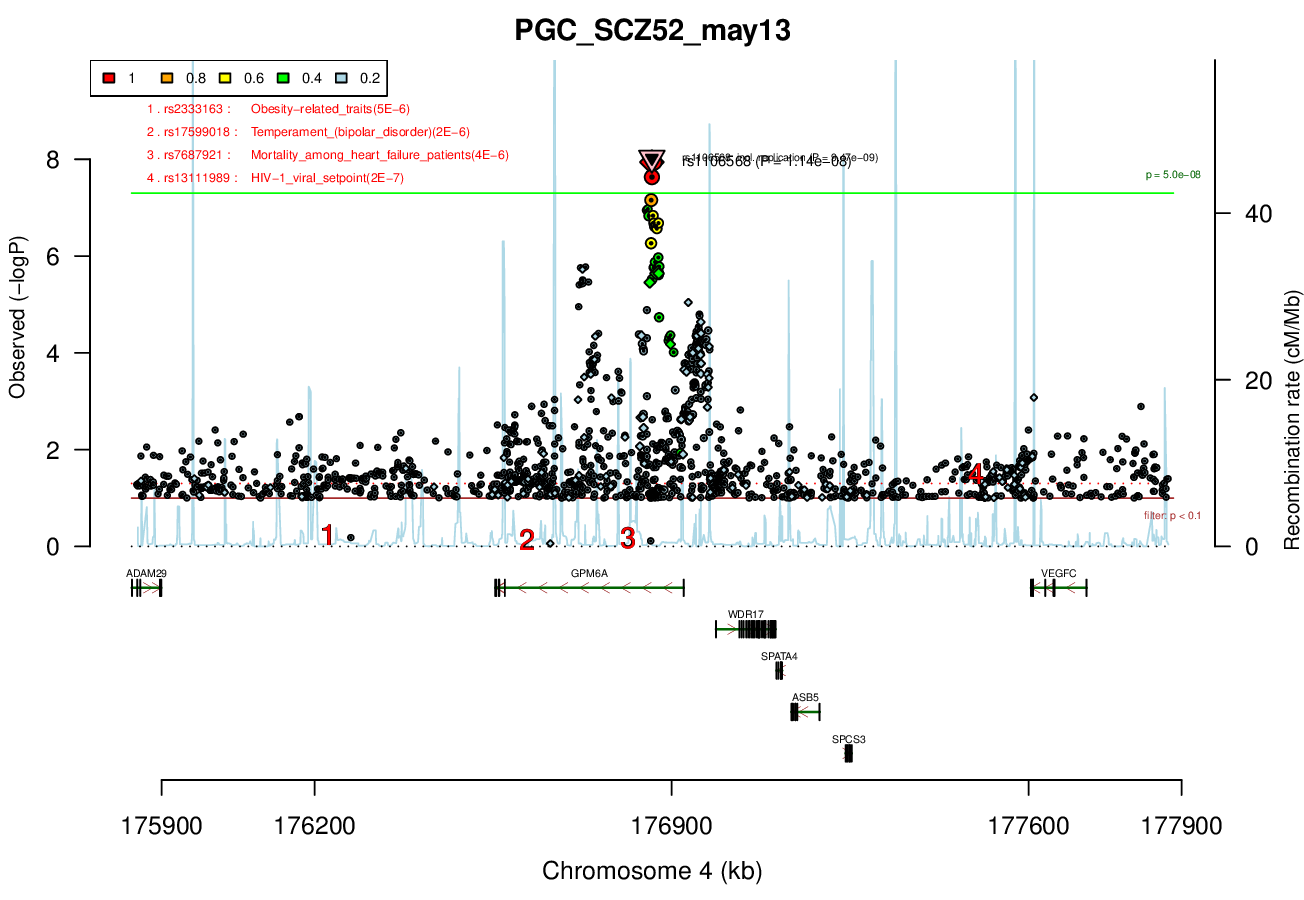
Distance On Each Side Of Locus In Above Plot
Ricopili Plot
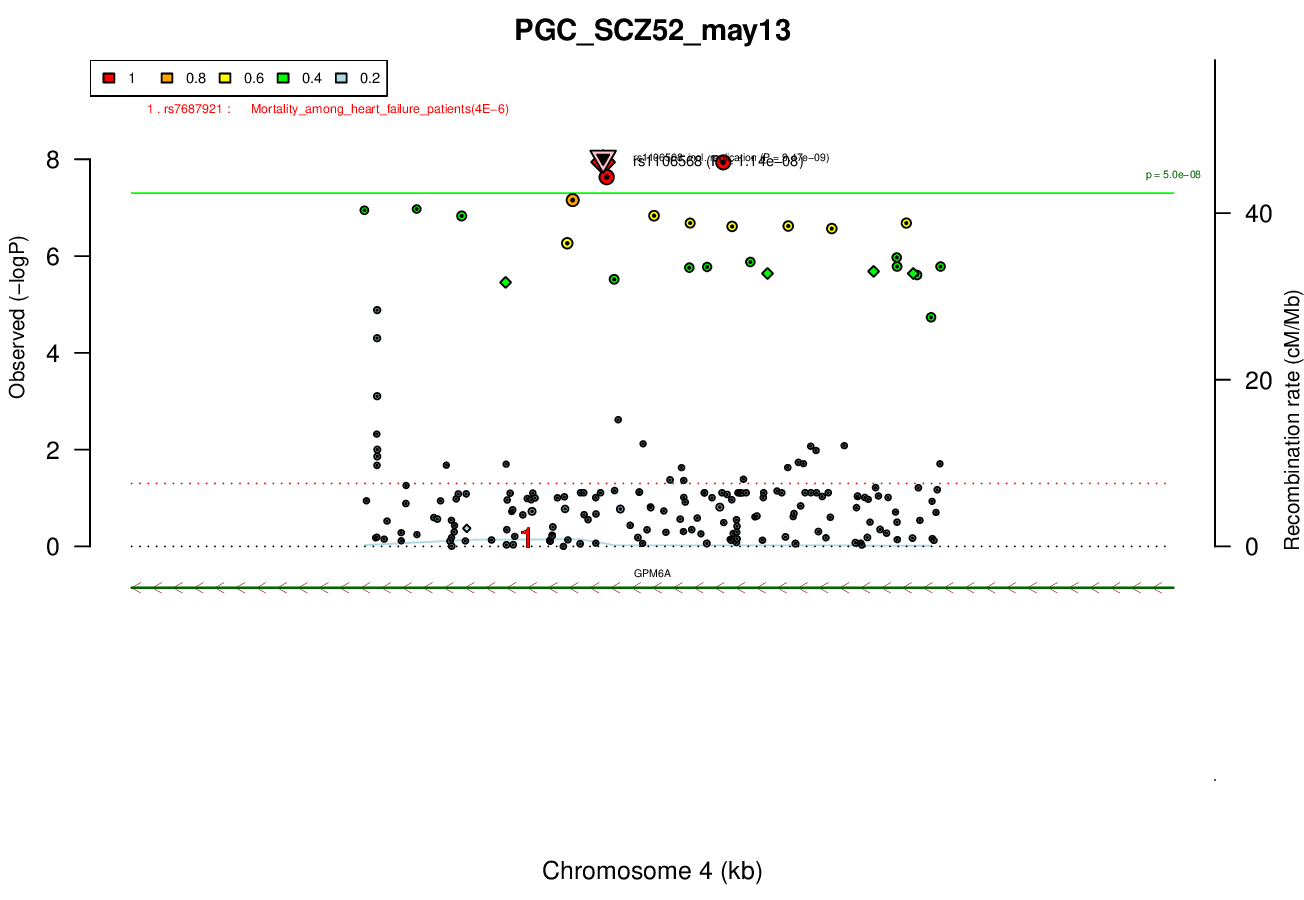
Gwas And Surrounding Region Pic

Gwas Region Pic
Sequence 1
Ensembl Gene Name
gpm6aaHarvard Allele
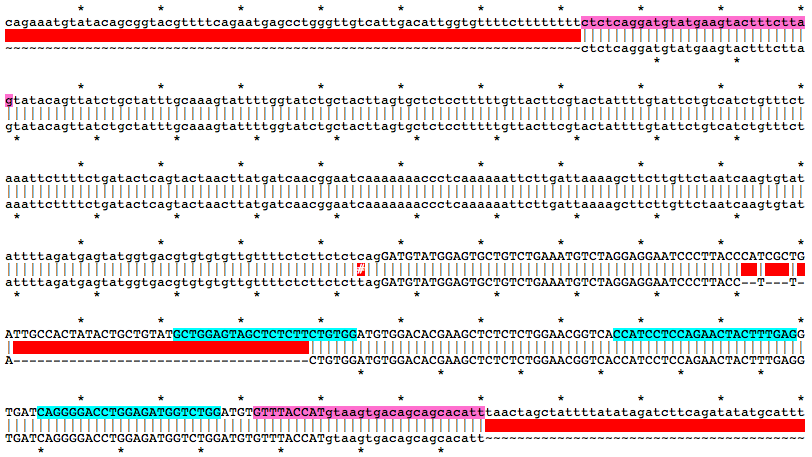
a275Mutation Area - WT DNA Sequence
ctctcaggatgtatgaagtactttcttagtatacagttatctgctatttgcaaagtattttggtatctgctacttagtgctctcctttttgttacttcgtactattttgtattctgtcatctgtttctaaattcttttctgatactcagtactaacttatgatcaacggaatcaaaaaaaccctcaaaaaattcttgattaaaagcttcttgttctaatcaagtgtatattttagatgagtatggtgacgtgtgtgttgttttctcttctctcagGATGTATGGAGTGCTGTCTGAAATGTCTAGGAGGAATCCCTTACCCATCGCTGATTGCCACTATACTGCTGTATGCTGGAGTAGCTCTCTTCTGTGGATGTGGACACGAAGCTCTCTCTGGAACGGTCACCATCCTCCAGAACTACTTTGAGGTGATCAGGGGACCTGGAGATGGTCTGGATGTGTTTACCATgtaagtgacagcagcacattMutation Area - Mutant DNA Sequence
ctctcaggatgtatgaagtactttcttagtatacagttatctgctatttgcaaagtattttggtatctgctacttagtgctctcctttttgttacttcgtactattttgtattctgtcatctgtttctaaattcttttctgatactcagtactaacttatgatcaacggaatcaaaaaaaccctcaaaaaattcttgattaaaagcttcttgttctaatcaagtgtatattttagatgagtatggtgacgtgtgtgttgttttctcttctcttagGATGTATGGAGTGCTGTCTGAAATGTCTAGGAGGAATCCCTTACCTTACTGTGGATGTGGACACGAAGCTCTCTCTGGAACGGTCACCATCCTCCAGAACTACTTTGAGGTGATCAGGGGACCTGGAGATGGTCTGGATGTGTTTACCATgtaagtgacagcagcacattGuide RNA Target Sites
CTCAAAGTAGTTCTGGAGGATGG
GCTGGAGTAGCTCTCTTCTGTGG
CAGGGGACCTGGAGATGGTCTGG
Genotyping Primers
f, ctctcaggatgtatgaagtactttcttag
r, aatgtgctgctgtcacttacATGGTAAAC
Alignment of WT and Mutant Sequences w/ gRNA Targets (cyan) and Genotyping Primers (pink) Shown on WT Sequence
Allele
43bpDWT Genotyping Size
488WT Protein Sequence
MEEDMDEGQTQKGCMECCLKCLGGIPYPSLIATILLYAGVALFCGCGHEALSGTVTILQN
YFEVIRGPGDGLDVFTIIDIIKYVIYGIASAFFVYGILLMVEGFFTSGAIKDLYGDFKIT
TCGRCVSAWFIMLTYIFMLAWLGVTAFTSLPVFIYFNIWTICQNTTVLQGASLCLDPRQF
GVVPIGEEKTLCVGSENFFKMCESNELDMTFHLFVCALAGAGAAVIAMIHYLMVLSANWA
YVKDACKMQKYEDCKSKEEQELHDIHSTRSKERLNAYT-
Mutant Protein Sequence
MEEDMDEGQTQKGCMECCLKCLGGIPYLTVDVDTKLSLERSPSSRTTLR-Mutant Genotyping Size
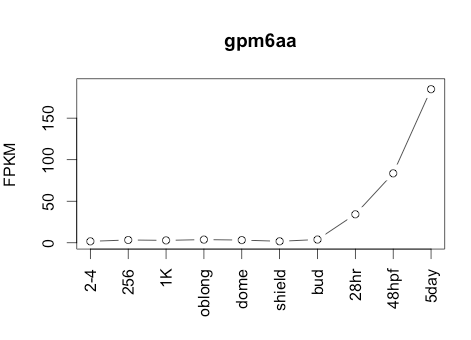
445Ribosome Profiling Development
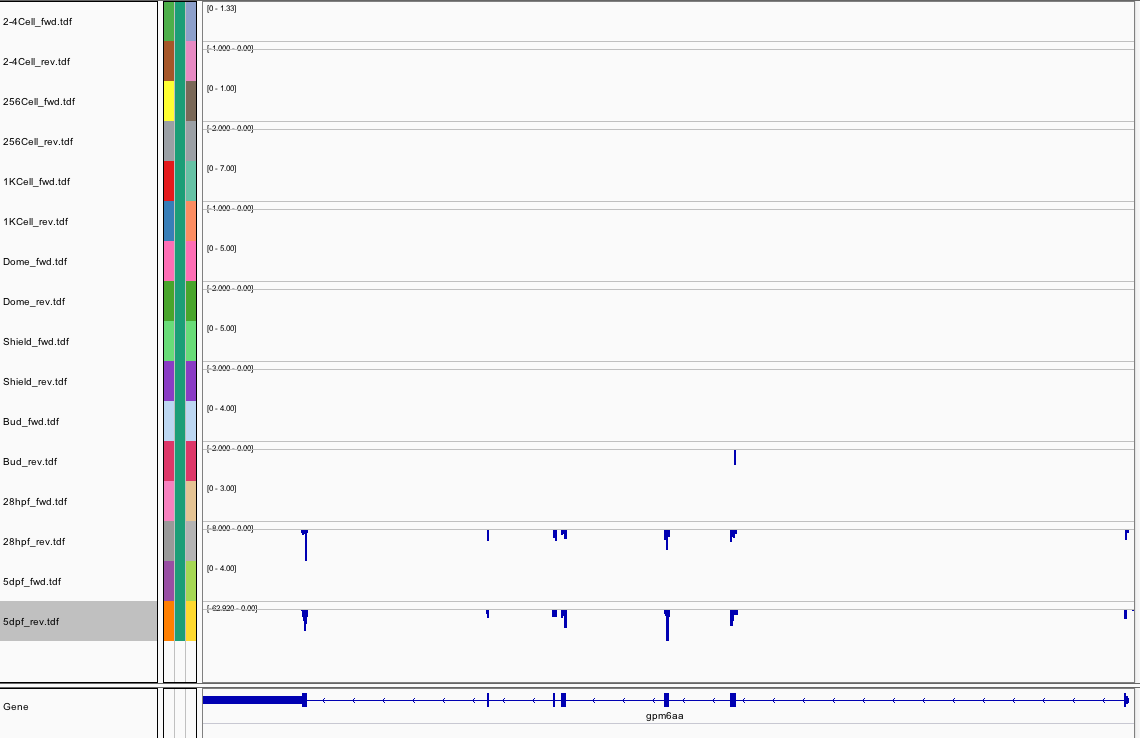
Transcript Plot
Sequence 2
Ensembl Gene Name
gpm6abHarvard Allele
a276Mutation Area - WT DNA Sequence
ttatcttctcagGATTGACATCTTCAAGTATGTGATCTATGGAGTGGCAGCAGCTTTTTTTGTCTATGGTATTTTGCTGATGGTGGAGGGATTTTTCACCACTGGTGCCATCCGGGATCTCTATGGAGACTTCAAGATCACCACCTGTGGGCGCTGTGTTAGTGCCTGGgtaagatcagacaaccatttaaaagaactaatgcgcagaagaaggggtcagttcacccaaaaatgtatattcagtcatcatttatgcacctttgctgttattttaatacaatatgcccttctgtctttttaaggatatttaattattgtgatataaatacagtttcacaagagttttatggcaatatttgatgattttctggggagtctaacattactcaggtagagaaattaaataatcacaatgcaaaaaaaaaaaaaaaaaaagcttttcttactttgctttgtcttgtttctagtctaattatcaaagaaattcttgtagcMutation Area - Mutant DNA Sequence
TTATCTTCTCAGGATTGACATCTTCAAGTATGTGATCTATGGAGTGGCAGCAGCTTTTTTTGTCTATGGTATTTTGCTGATGGTGGAGGGATTTTTCACCGTGCCTGGGTAAGATCAGACAACCatttaaaagaactaatgcgcagaagaaggggtcagttcacccaaaaatgtatattcagtcatcatttatgcacctttgctgttattttaatacaatatgcccttctgtctttttaaggatatttaattattgtgatataaatacagtttcacaagagttttatggcaatatttgatgattttctggggagtctaacattactcaggtagagaaattaaataatcacaatgcaaaaaaaaaaaaaaaaaaagcttttcttactttgctttgtcttgtttctagtctaattatcaaagaaattcttgtagcGuide RNA Target Sites
GAGATCCCGGATGGCACCAGTGGGenotyping Primers
f, ttatcttctcagGATTGACATCTTCAAGTATG
r, gctacaagaatttctttgataattagactagaaacaagac
Alignment of WT and Mutant Sequences w/ gRNA Targets (cyan) and Genotyping Primers (pink) Shown on WT Sequence
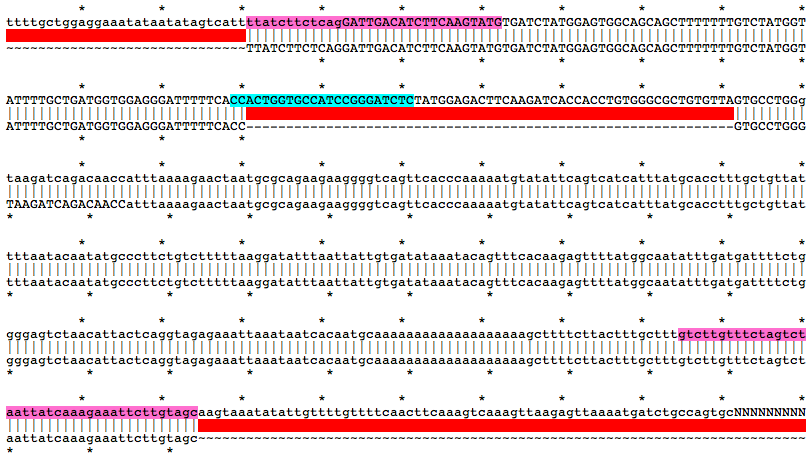
Allele
61bpDWT Genotyping Size
494WT Protein Sequence
MGCSECCLKCLSGIPYASLIATILLYAGVALFCGCGHEALSG
TVTILQTYFDVVRSPVDALDVFTMIDIFKYVIYGVAAAFFVYGILLMVEGFFTTGAIRDL
YGDFKITTCGRCVSAWFIMLTYIFMLAWLGVTAFTSLPVFMYFNIWNTCQNMTLLEGSLS
SANSANLCFDLRQYGIVSIGEEKRLCADNENFKKMCESNELDLTFHLFVCALAGAGAAVI
AMVHYLMVLSANWAYVKDACRMQKYEDIKSKEEQELHDIHSTRSKERLNAYT-
Mutant Protein Sequence
MGCSECCLKCLSGIPYASLIATILLYAGVALFCGCGHEALSG
TVTILQTYFDVVRSPVDALDVFTMIDIFKYVIYGVAAAFFVYGI
LLMVEGFFTVPG-
In Situ Probe Sequence
ATCCCGTATGCCTCTCTGATCGCCACTATCCTGCTGTACGCCGGTGTTGCTCTCTTCTGTGGCTGTGGACATGAGGCTCTTTCTGGCACCGTCACCATCCTGCAGACCTACTTTGATGTCGTCCGGTCGCCTGTTGATGCCCTGGATGTCTTTACCATGATTGACATCTTCAAGTATGTGATCTATGGAGTGGCAGCAGCTTTTTTTGTCTATGGTATTTTGCTGATGGTGGAGGGATTTTTCACCACTGGTGCCATCCGGGATCTCTATGGAGACTTCAAGATCACCACCTGTGGGCGCTGTGTTAGTGCCTGGTTCATCATGCTTACTTATATCTTCATGTTGGCCTGGCTGGGAGTCACAGCCTTCACCTCTCTGCCGGTCTTCATGTACTTCAACATCTGGAACACTTGTCAGAACATGACTCTGCTGGAGGGTTCTCTCTCTTCAGCGAACAGCGCCAACCTTTGCTTTGACCTGCGTCAGTACGGAATCGTGTCCATCGGTGAAGAGAAAAGACTGTGTGCCGATAATGAAAACTTCAAGAAGATGTGTGAATCTAATGAGCTGGATCTGACGTTCCACTTGTTTGTCTGCGCTCTTGCGGGGGCTGGAGCGGCGGTTATCGCCATGGTGGTGGCCGTGTCTGTCCTCATCAATAACCACGCCAGTCTGACATCTAAGAACGCAGGGAGGTACTMutant Genotyping Size
433Ribosome Profiling Development
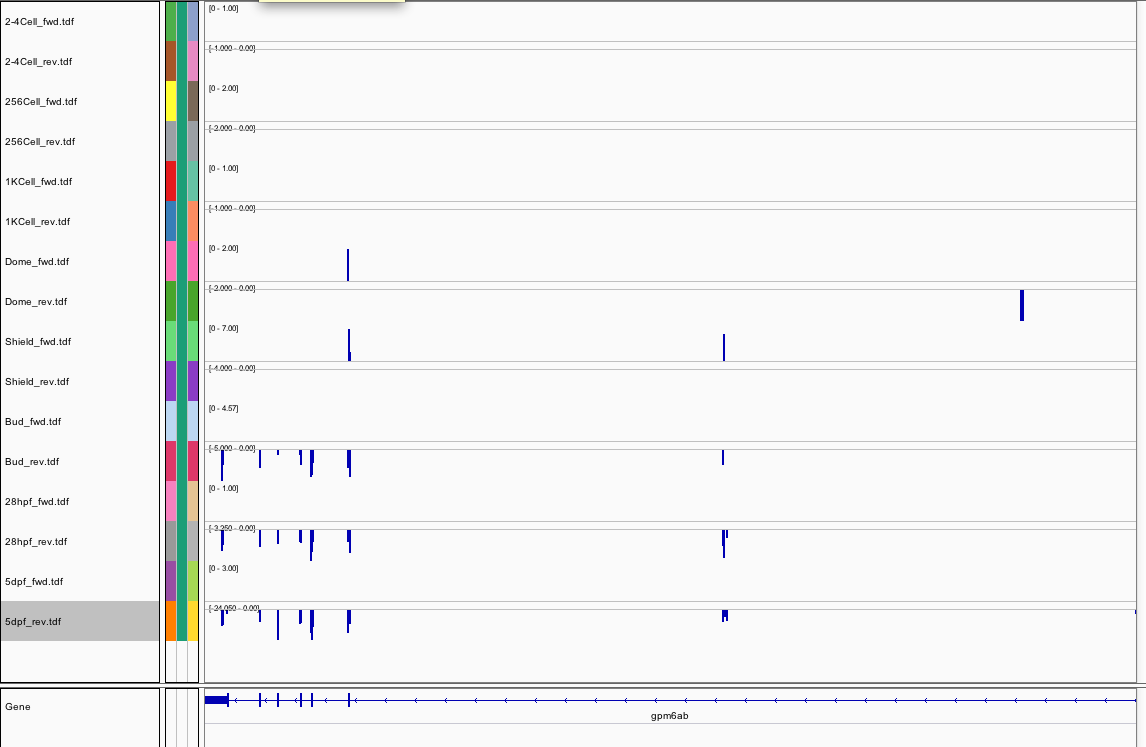
Transcript Plot
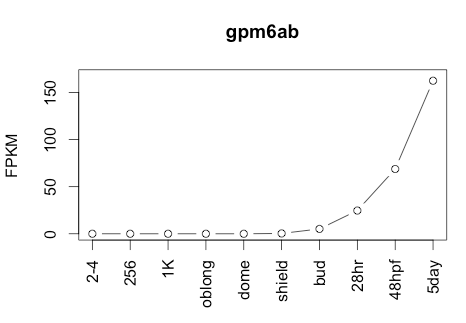
Behavior Data
Behavior Data Summary
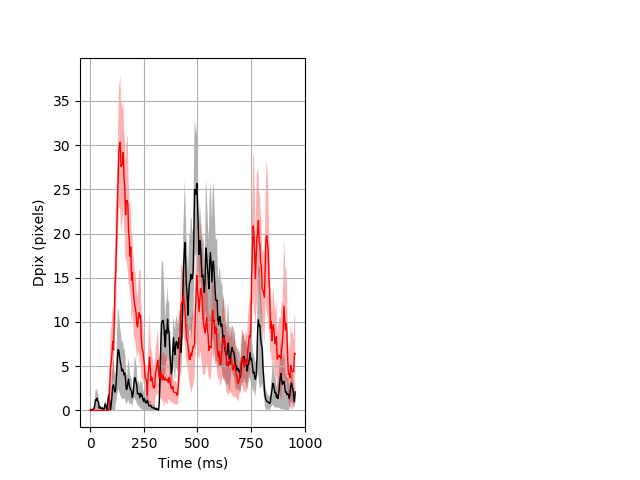
NoneBehavior Data Description 1
Dark flash block 1 start Merged section Pvalue = 0.0303 7 hethom vs 7 homhom ribgraph_mean_ribbon_fullboutdata_dpix_a0darkflash103.pngBehavior Data Graph 1
Behavior Data Description 2
Day all prepulse tap Merged section Pvalue = 0.0233 7 hethet vs 7 homhom ribgraph_mean_ribbon_fullboutdata_dpix_dayprepulseinhibition100d.pngBehavior Data Graph 2
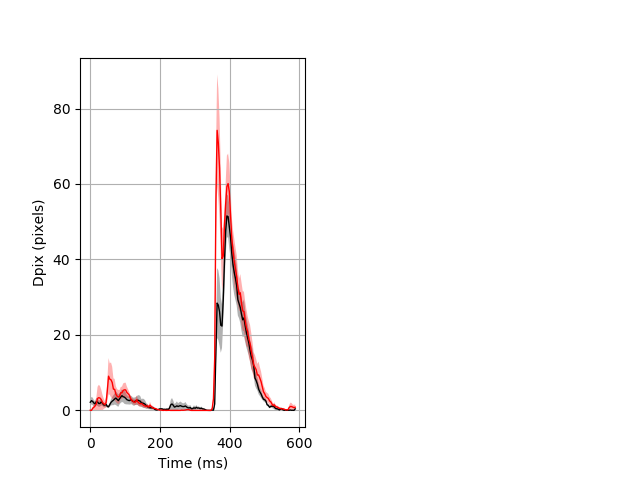
Behavior Data Description 3
Features of movement entire protocol Graph Pvalue = 0.0476 Merged section Pvalue = not significant 7 hethet vs 7 homhom ribgraph_mean_ribbonbout_aveboutdisp_10min_combo.pngBehavior Data Graph 3
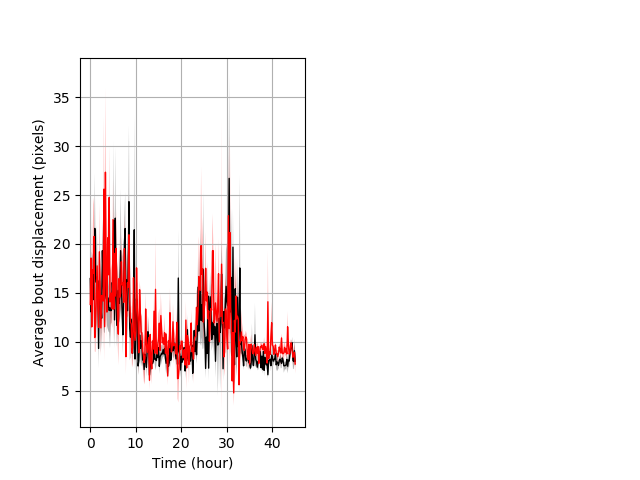
Behavior Data Description 4
Frequency of movement entire protocol Graph Pvalue = not significant Merged section Pvalue = not significant 6 wthet vs 7 homhom ribgraph_mean_ribbonbout_dpixnumberofbouts_10min_combo.pngBehavior Data Graph 4
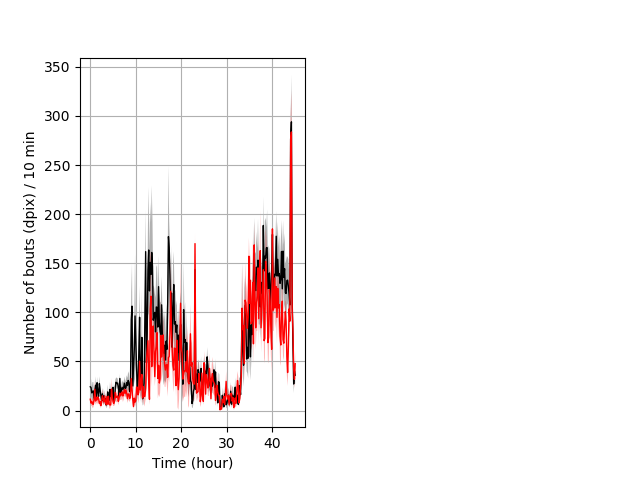
Behavior Data Description 5
Location in well entire protocol Graph Pvalue = 0.0321 Merged section Pvalue = 0.027 might not repeat 7 hethet vs 6 wthom ribgraph_mean_ribbonbout_averhofrac_10min_combo.pngBehavior Data Graph 5
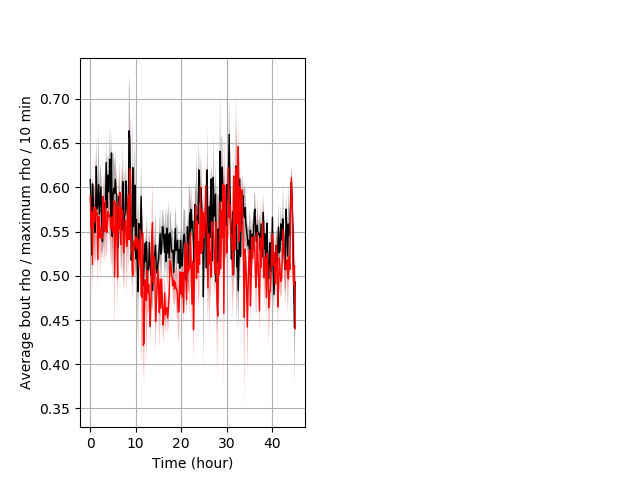
Behavior Data Description 6
Frequency of movement Graph Pvalue = 0.001 6 wthet vs 7 homhom ribgraph_mean_ribbon_distsecper_min_day1mornstim.pngBehavior Data Graph 6
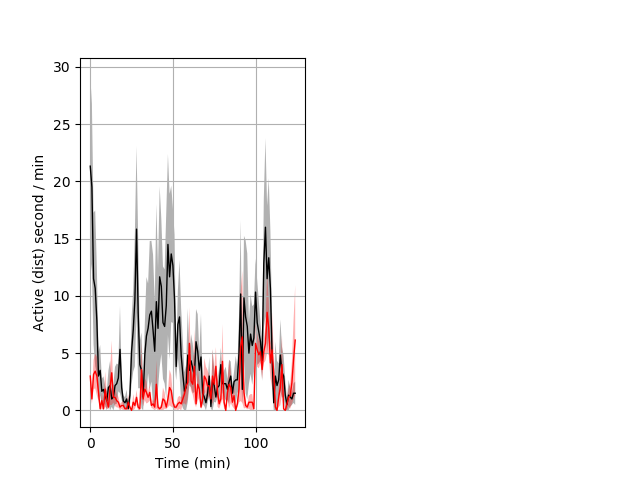
Behavior Data Description 7
Location in well Graph Pvalue = 0.001 10 homhet vs 7 homhom ribgraph_mean_ribbonbout_averhofrac_10min_day2morning.pngBehavior Data Graph 7
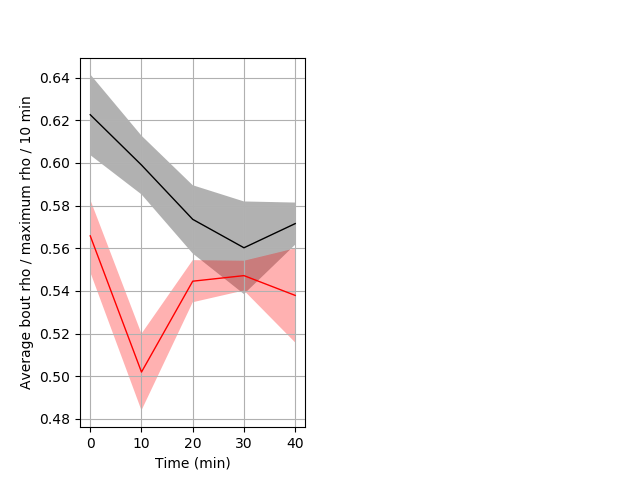
Behavior Data Description 8
Location in well Graph Pvalue = 0.004 7 hethet vs 6 wthom ribgraph_mean_ribbonbout_interboutaverhofrac_10min_day1taps.pngBehavior Data Graph 8
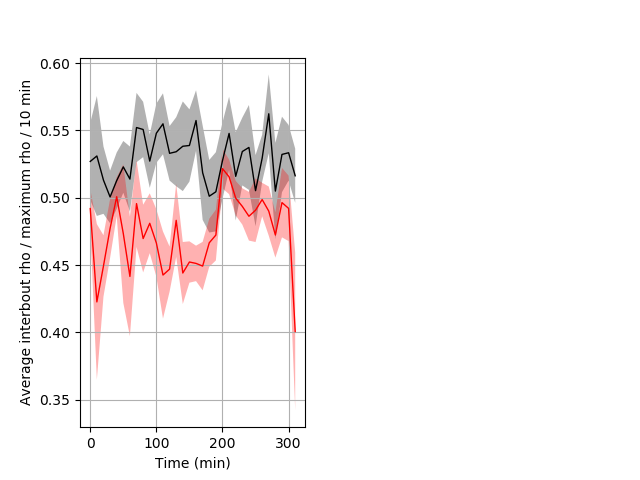
Behavior Data Description 9
PPI tap Graph Pvalue = 0.0164 15 homhet vs 11 homhom boxgraph_pd__ribgraph_mean_ribbon_totaldistanceresponse_dist_nightprepulseinhibition100b.pngBehavior Data Graph 9
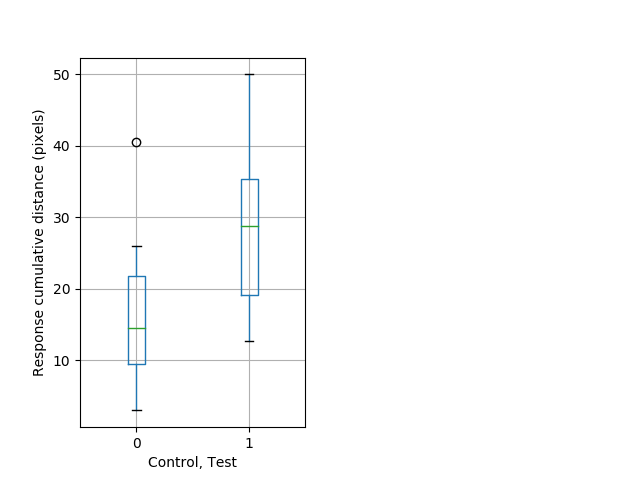
Behavior Data Description 10
Strong tap Graph Pvalue = 0.00415 17 hethom vs 11 homhom boxgraph_pd__smallmovesribgraph_mean_ribbon_latencyresponse_dpix_nightprepulseinhibition102.pngBehavior Data Graph 10
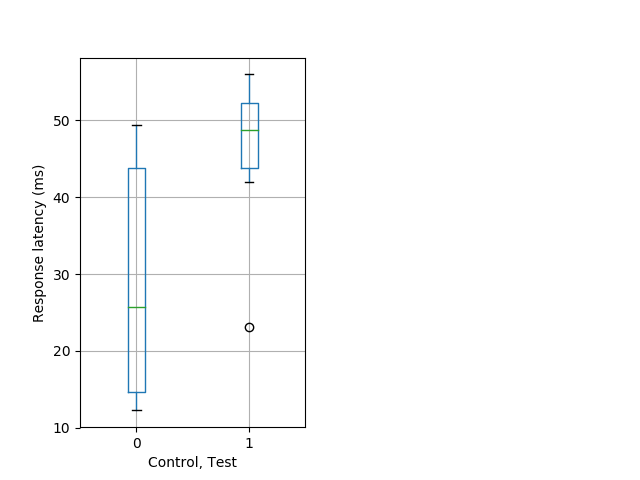
Behavior Data Description 11
Tap habituation Graph Pvalue = 0.0013 17 hethom vs 11 homhom boxgraph_pd__ribgraph_mean_ribbon_fullboutdatamax_dpix_cdaytaphab102.pngBehavior Data Graph 11
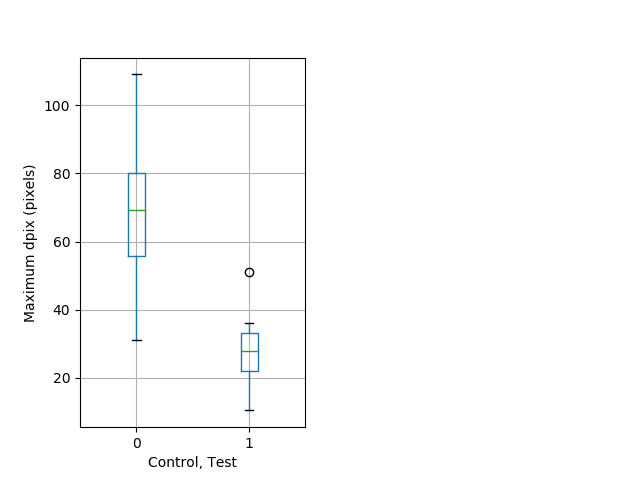
Behavior Data Description 12
Weak tap Graph Pvalue = 0.00877 17 hethom vs 11 homhom boxgraph_pd__ribgraph_mean_ribbon_timeresponse_dpix_nightprepulseinhibition101.pngBehavior Data Graph 12
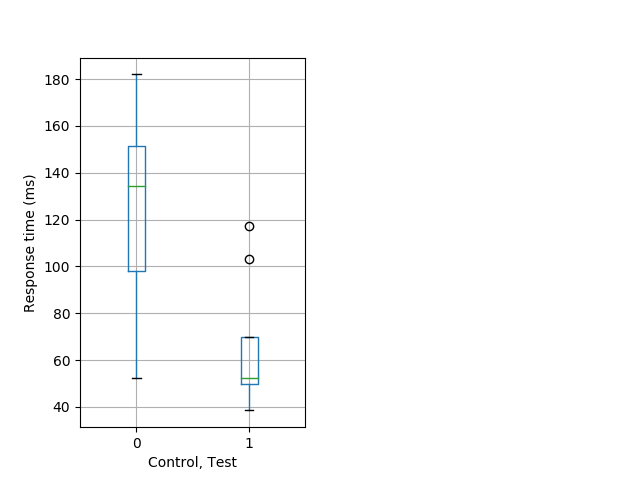
Behavior Data Description 13
NoneBehavior Data Graph 13
NoneBehavior Data Description 14
NoneBehavior Data Graph 14
None