Locus Rank
44Gene
Id
188Gene Name
GRM3Duplicated
FalseMaternal
FalseGene Not Within Locus (Nearby
FalseGene Description
L-glutamate is the major excitatory neurotransmitter in the central nervous system and activates both ionotropic and metabotropic glutamate receptors. Glutamatergic neurotransmission is involved in most aspects of normal brain function and can be perturbed in many neuropathologic conditions. The metabotropic glutamate receptors are a family of G protein-coupled receptors, that have been divided into 3 groups on the basis of sequence homology, putative signal transduction mechanisms, and pharmacologic properties. Group I includes GRM1 and GRM5 and these receptors have been shown to activate phospholipase C. Group II includes GRM2 and GRM3 while Group III includes GRM4, GRM6, GRM7 and GRM8. Group II and III receptors are linked to the inhibition of the cyclic AMP cascade but differ in their agonist selectivities. Mouse Phenotype
Additional Image
Allen Brain
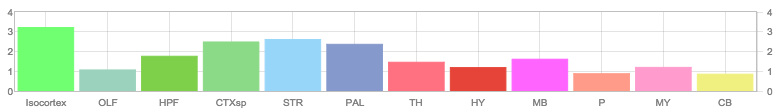
Allen Regions
Zfin In Situ
Zfin Brain Areas
caudal tuberculum dorsal region, caudal tuberculum ventral region, caudal tuberculum ventro-lateral region, corpus cerebelli, dorsal telencephalon, fourth ventricle, habenula, hypothalamus lateral recess, hypothalamus posterior region, inferior olive, intermediate hypothalamus, retinal ganglion cell layer, retinal inner nuclear layer, superior raphe nucleus, tectal ventricle, telencephalic ventricle, valvula cerebelliPublished Zebrafish Pehnotype
Not In Allen Brain
FalseZFIN Link
http://zfin.org/ZDB-GENE-061009-13Allen Link
NoneMore Additional Images
Papers
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3681425/
http://www.ncbi.nlm.nih.gov/pubmed/25179095
Locus Rank
Locus Rank
44Genes in Region
GRM3Associated Snps
rs12704290GWAS Region (hg19)
chr7:86403226-86459326Genes Skipping
Ricopili Plot Surrounding Area
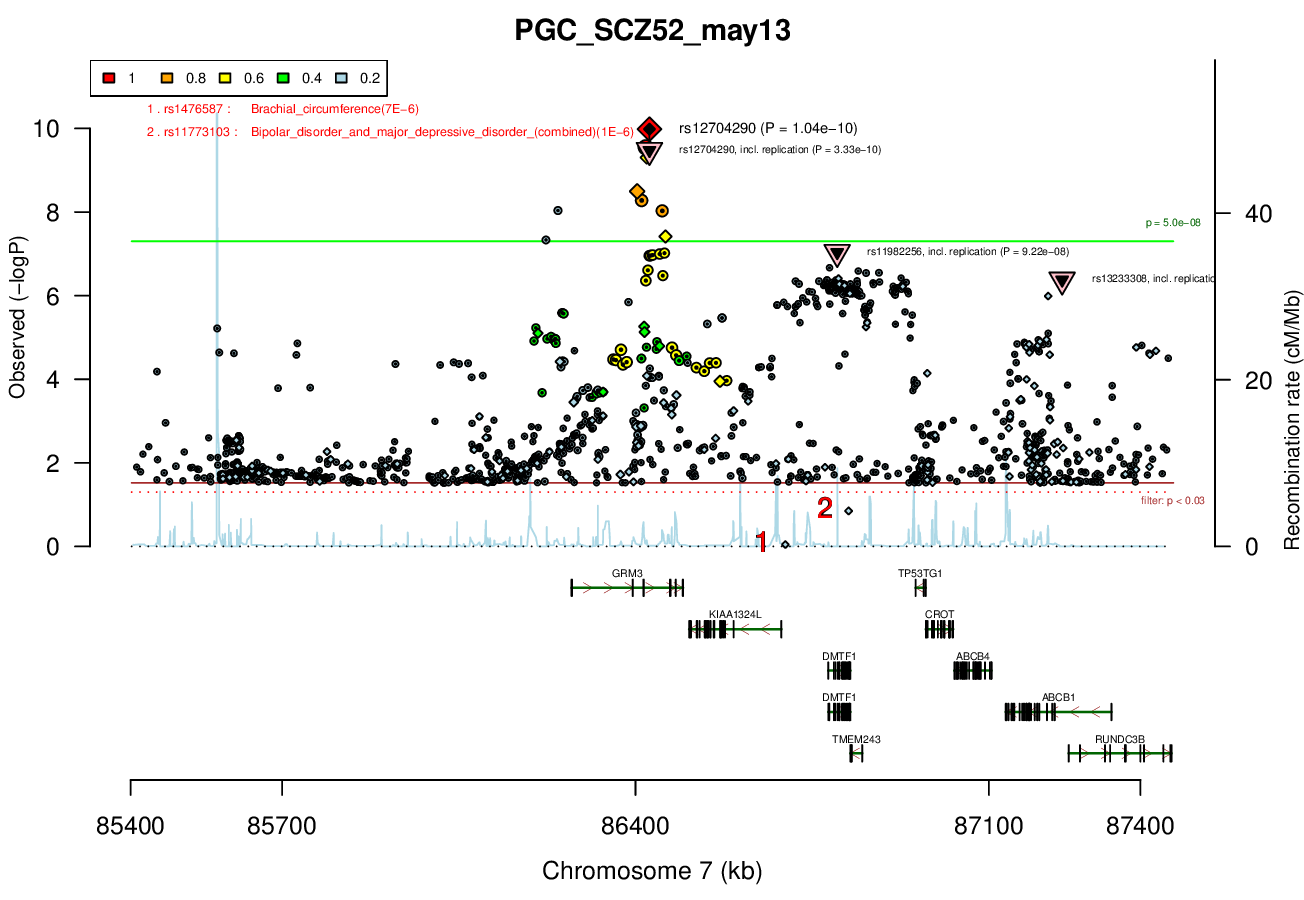
Distance On Each Side Of Locus In Above Plot
1 MBRicopili Plot
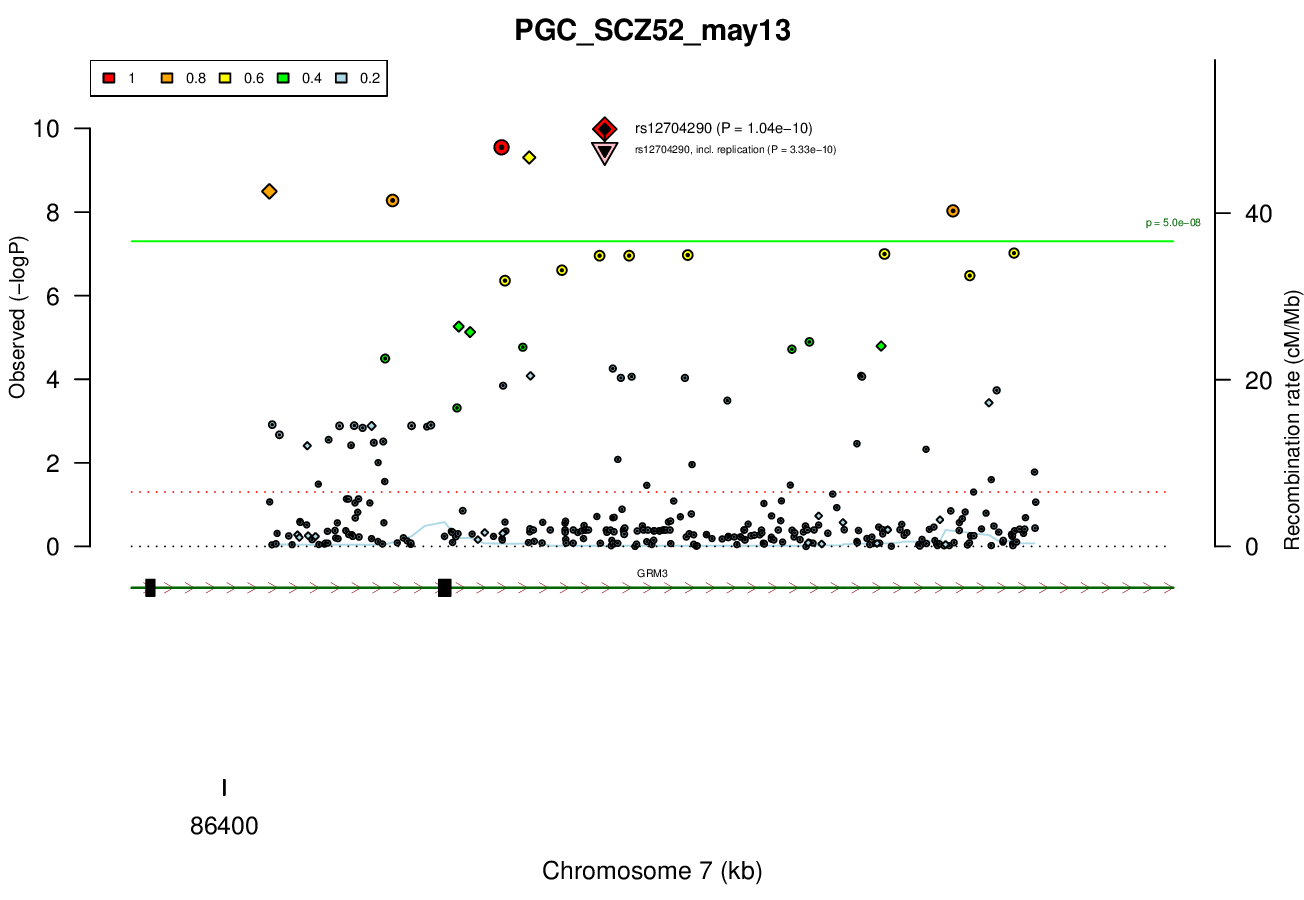
Gwas And Surrounding Region Pic
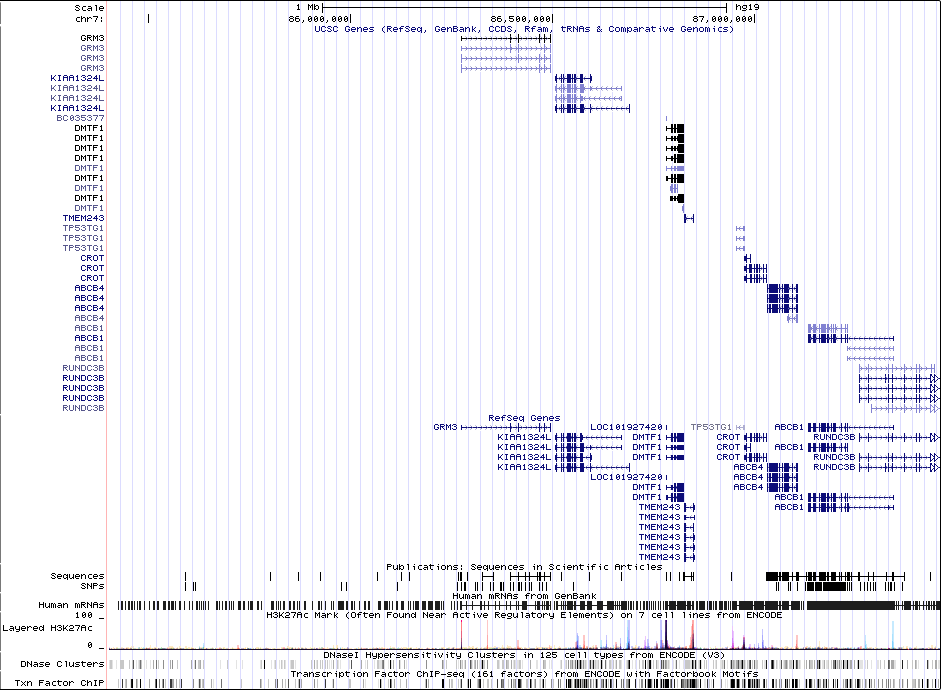
Gwas Region Pic
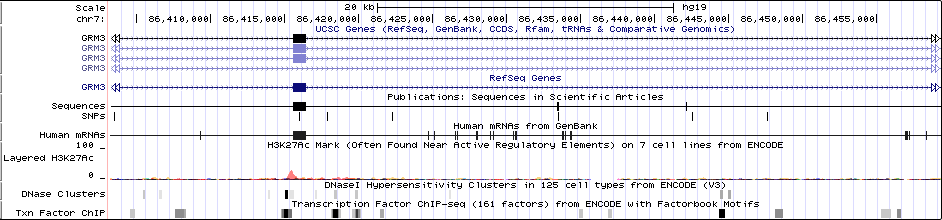
Sequence 1
Ensembl Gene Name
grm3Harvard Allele
a281Mutation Area - WT DNA Sequence
ACCAGGCCAAGGCCATGGCGGAGATCTTACGGGCCTTCAACTGGACCTACGTTTCCACCGTGGCCTCAGAGGGAGACTATGGAGAAACAGGAATCGAAGCCTTCGAGCAACAAGCGCGTTTGAGAAACATCTGCATCGCTACTTCGGAAAAAGTAGGACGATCTAGCGCAAAGCGTTCTTCATATGAGGCCGTGGTCCGCCAGCTCCTCCAGAAGCCCACAGCTCGCGTCGCTGTTCTTTTTCTACGTAGCGACGACGCAAGAGAACTAATCGCCGCTGCCGCCCGCCTCAACGCATCTTTCCTCTGGGTGGCCAGCGACGGATGGGGGGCGCAAGAGAGCATCGTCAAAGGGAACGAATTCACAGCAGATGGTGCAATCACGCTGGAGCTAGCGGCTCATCCCATACCAGATTTCAACCGTTACTTCCAGACCCTCACGCCCCAAATTAACCATCGCAACCCCTGGTTCAAAGACTTCTGGGAGCAAAAGTTTCAGTGTTCATTGGGTGGGACGTCAGCAACAGGGGCAGGAACCCCAAAGCCACCTTGTGACCAGGAACTGGTTATCGACAAGTCCAATTTTGAGCCAGAATCAAAGATTATGTTTGTGGTAAATGCAGTGTATGCCATGGCGCATGCTTTGCATCGCATGCAACGGACTTTGTGTGCCAACACTACACGTTTGTGTGATGCCATGCGCAGTCTTGMutation Area - Mutant DNA Sequence
ACCAGGCCAAGGCCATGGCGGAGATCTTACGGGCCTTCAACTGGACCTACGTTTCCACCGTGGCCTCAGAGGGAGACTATGGAGAAACAGGAATCGAAGCCTTCGAGCAACAAGCGCGTTTGAGAAACATCTGCATCGCTACTTCGGAAAAAGTAGGACGATCGAGCGCAAAGCGTTCTTCATATGAGGCCGTGGTCCGCCAGCTCCTCCAGAAGCCCACAGCTCGCGTCGCTGTTCTTTTTCTACGTAGCGACGACGCAAGAGAACTAATCGCCGCTGCCGCCCGCCTGTGACCAGGAACTGGTTATCGACAAGTCCAATTTTGAGCCAGAATCAAAGATCATGTTTGTGGTAAATGCGGTGTATGCCATGGCGCATGCTTTGCATCGCATGCAACGGACTTTGTGTGCCAACACTACACGTTTGTGTGATGCCATGCGCAGTCTTGGuide RNA Target Sites
GCGTTCTTCATATGAGGCCGTGG
AGAGGAAAGATGCGTTGAGGCGG
TGCAATCACGCTGGAGCTAGCGG
ACCAGTTCCTGGTCACAAGGTGG
Genotyping Primers
f, ACCAGGCCAAGGCCATGGCGG
r, CAAGACTGCGCATGGCATCACAC
Alignment of WT and Mutant Sequences w/ gRNA Targets (cyan) and Genotyping Primers (pink) Shown on WT Sequence
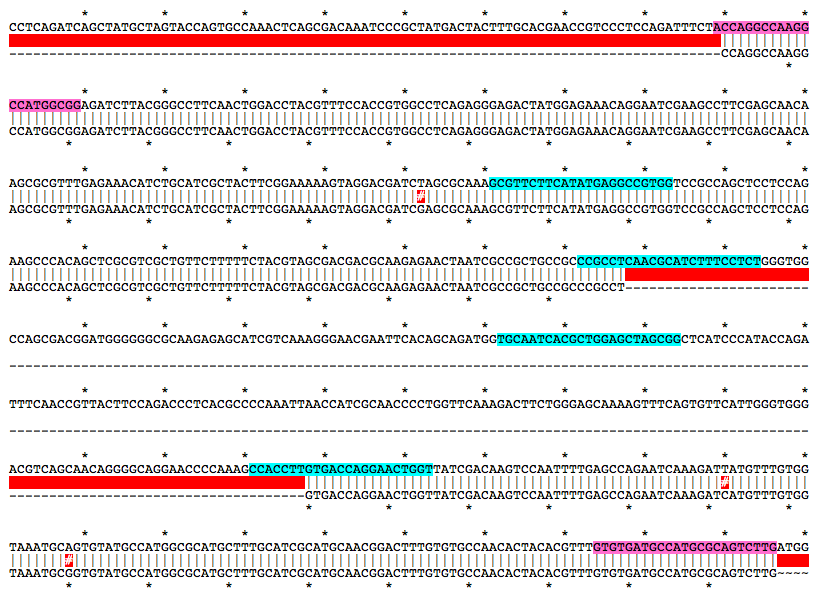
Allele
260bpDWT Genotyping Size
708WT Protein Sequence
MWSAVRALLLLSAGLLQSRGDSPRKEIKIDGDLVLGGLFPIHEKGMGMDECGRINEDRGI
QRLEAMLFAIDQINQDVALLPGVVLGVHILDTCSRDTYALEQALEFVRASLTKVDDTEFI
CPDGSYALQEDSPLAIAGVIGGSFSSVSIQVANLLRLFQIPQISYASTSAKLSDKSRYDY
FARTVPPDFYQAKAMAEILRAFNWTYVSTVASEGDYGETGIEAFEQQARLRNICIATSEK
VGRSSAKRSSYEAVVRQLLQKPTARVAVLFLRSDDARELIAAAARLNASFLWVASDGWGA
QESIVKGNEFTADGAITLELAAHPIPDFNRYFQTLTPQINHRNPWFKDFWEQKFQCSLGG
TSATGAGTPKPPCDQELVIDKSNFEPESKIMFVVNAVYAMAHALHRMQRTLCANTTRLCD
AMRSLDGRKLYRDFLLHVNFRAPFSPAGLESQVKFDAYGDGVGRYNIFNYQSVPGSDKFA
YVQVGEWAESLSLNEGLIGWPRGADVPTSQCSDPCAPNEMKKMQAGEYCCWICTPCEPYE
YLPDEFTCMPCAPGQWPRPDLTGCYDLPEDYIMWEDAWAIGPISIACVGFICTLMVFIVF
IRHNDTPLVKASGRELCYILLLGVFMSYVMTFIFIAKPSPIVCTLRRLGLGTSFAVCYSA
LLTKTNRIARIFSGVKEGGAQRPRFISPSSQVFICLSLISVQLLLVSVWLLVEVPGTRRF
TTPEKRQTVILKCNVRDSSMLMSLSYDVVLVVLCTVYAFKTRKCPENFNEAKFIGFTMYT
TCIIWLAFLPIFYVTSSDYRVQTTTMCISVSLSGFVVLGCMFAPKVHIIMFQPQKNVTSH
RLNMNRFSVSGPATSYASHASVSAHYVPTVCNGREIVDSTTSSL-
Mutant Protein Sequence
MWSAVRALLLLSAGLLQSRGDSPRKEIKIDGDLVLGGLFPIHEKGMGMDECGRINEDRGI
QRLEAMLFAIDQINQDVALLPGVVLGVHILDTCSRDTYALEQALEFVRASLTKVDDTEFI
CPDGSYALQEDSPLAIAGVIGGSFSSVSIQVANLLRLFQIPQISYASTSAKLSDKSRYDY
FARTVPPDFYQAKAMAEILRAFNWTYVSTVASEGDYGETGIEAFEQQARLRNICIATSE
KVGRSSAKRSSYEAVVRQLLQKPTARVAVLFLRSDDARELIAAAARL-
Mutant Genotyping Size
448Ribosome Profiling Development
Behavior Data
Behavior Data Summary
NoneBehavior Data Description 1
Dark flash block 1 start Merged section Pvalue = not significant 29 wt vs 18 hom ribgraph_mean_ribbon_fullboutdata_dpix_a0darkflash103.pngBehavior Data Graph 1
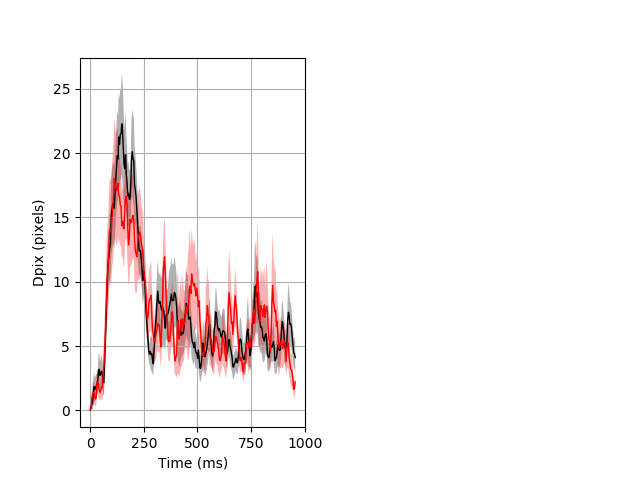
Behavior Data Description 2
Day all prepulse tap Merged section Pvalue = not significant 29 wt vs 18 hom ribgraph_mean_ribbon_fullboutdata_dpix_dayprepulseinhibition100d.pngBehavior Data Graph 2
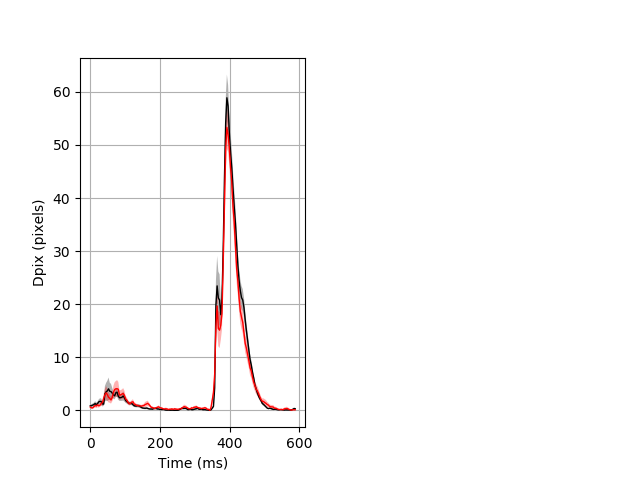
Behavior Data Description 3
Features of movement entire protocol Graph Pvalue = not significant Merged section Pvalue = not significant 29 wt vs 18 hom ribgraph_mean_ribbonbout_aveboutdisp_10min_combo.pngBehavior Data Graph 3
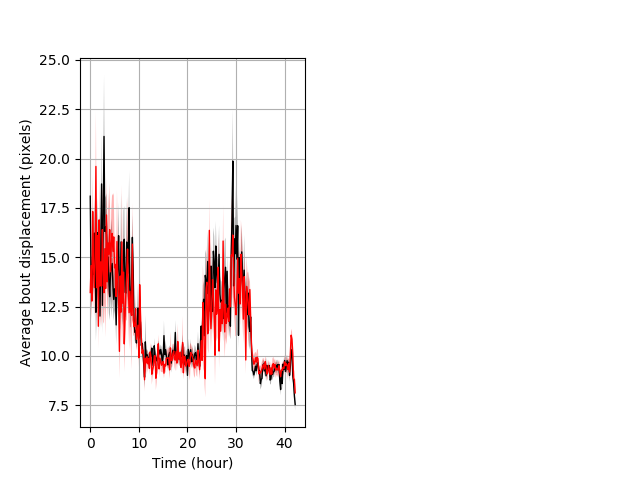
Behavior Data Description 4
Frequency of movement entire protocol Graph Pvalue = not significant Merged section Pvalue = not significant 29 wt vs 18 hom ribgraph_mean_ribbonbout_dpixnumberofbouts_10min_combo.pngBehavior Data Graph 4

Behavior Data Description 5
Location in well entire protocol Graph Pvalue = not significant Merged section Pvalue = not significant 29 wt vs 18 hom ribgraph_mean_ribbonbout_averhofrac_10min_combo.pngBehavior Data Graph 5
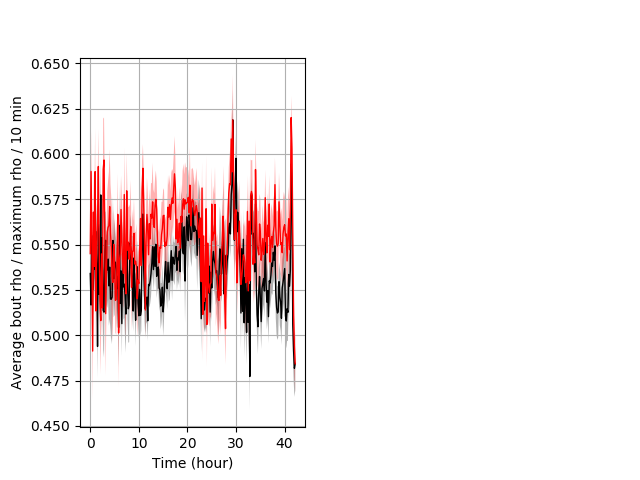
Behavior Data Description 6
Features of movement Graph Pvalue = 0.008 22 het vs 20 hom ribgraph_mean_ribbonbout_aveboutcumdpix_min_day1day.pngBehavior Data Graph 6
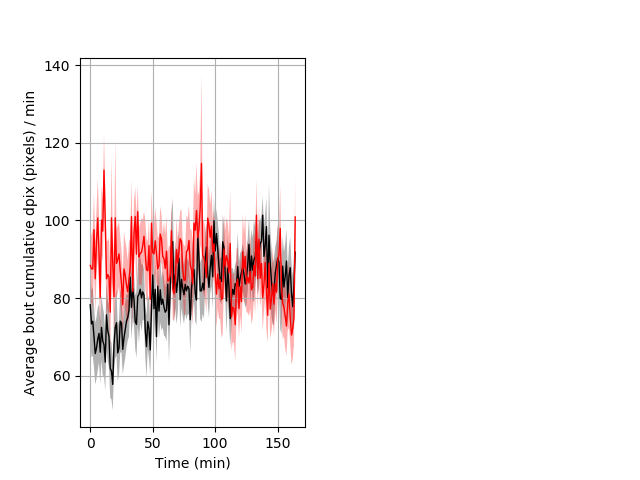
Behavior Data Description 7
Strong tap Graph Pvalue = 0.000942 35 het vs 18 hom boxgraph_pd__bigmovesribgraph_mean_ribbon_peakspeed_adaytappostbdaytappre102.pngBehavior Data Graph 7
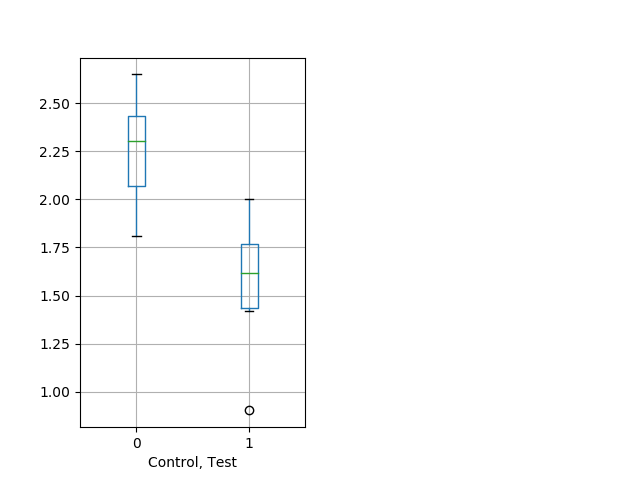
Behavior Data Description 8
NoneBehavior Data Graph 8
NoneBehavior Data Description 9
NoneBehavior Data Graph 9
NoneBehavior Data Description 10
NoneBehavior Data Graph 10
NoneBehavior Data Description 11
NoneBehavior Data Graph 11
NoneBehavior Data Description 12
NoneBehavior Data Graph 12
NoneBehavior Data Description 13
NoneBehavior Data Graph 13
NoneBehavior Data Description 14
NoneBehavior Data Graph 14
None