Locus Rank
58Gene
Id
276Gene Name
GTDC1Duplicated
FalseMaternal
FalseGene Not Within Locus (Nearby
TrueGene Description
Mouse Phenotype
Additional Image
Allen Brain
Allen Regions
Zfin In Situ
Zfin Brain Areas
Published Zebrafish Pehnotype
Not In Allen Brain
FalseZFIN Link
NoneAllen Link
NoneMore Additional Images
Papers
Locus Rank
Locus Rank
58Genes in Region
No genes within locus (gene desert). Both ZEB2 and GTDC1 are nearby.Associated Snps
GWAS Region (hg19)
chr2:146416922-146441832Genes Skipping
ZEB2, but it is a likely candidate and should be tested.Ricopili Plot Surrounding Area
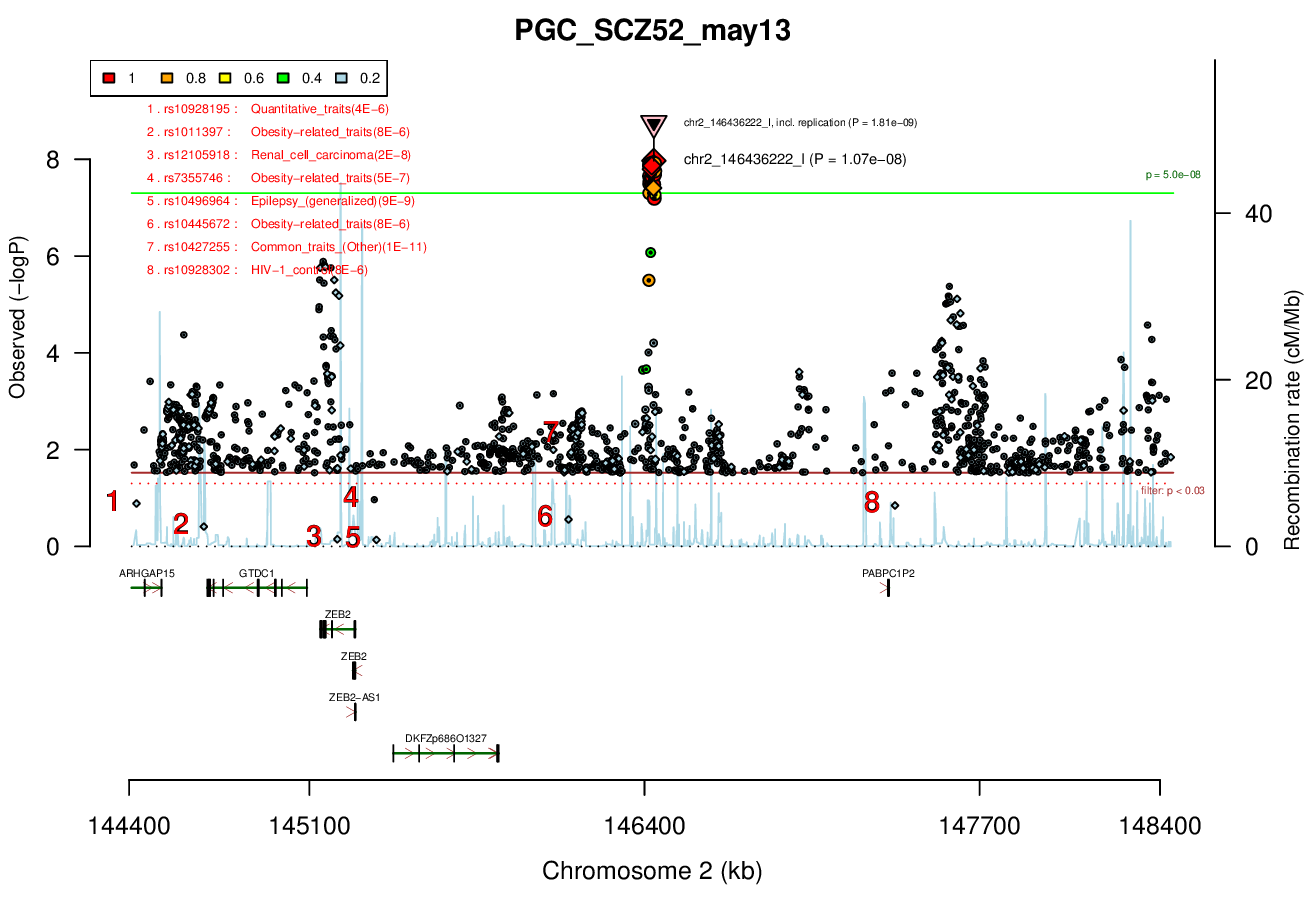
Distance On Each Side Of Locus In Above Plot
Ricopili Plot
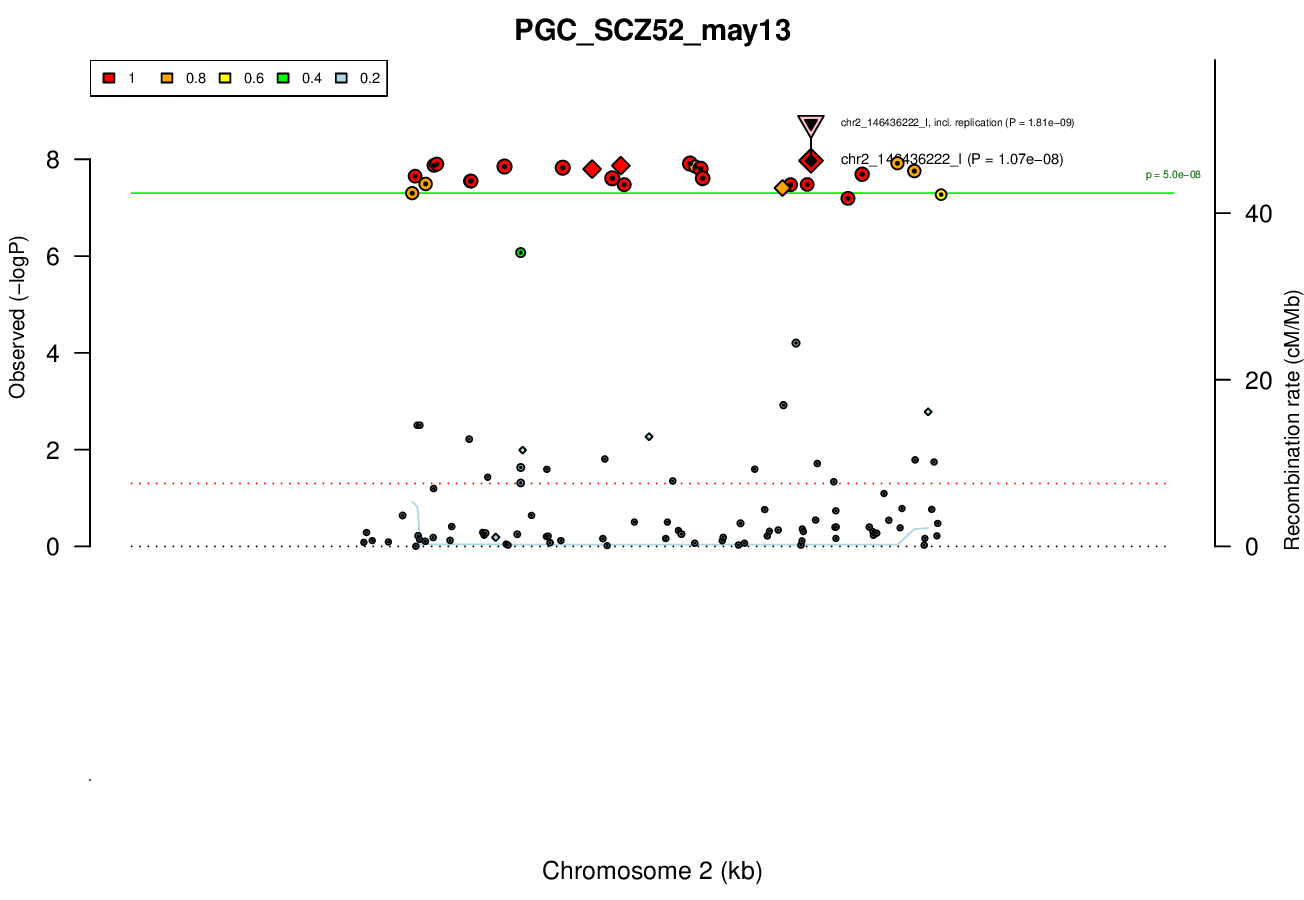
Gwas And Surrounding Region Pic
Gwas Region Pic
Sequence 1
Ensembl Gene Name
gtdc1Harvard Allele
a374Mutation Area - WT DNA Sequence
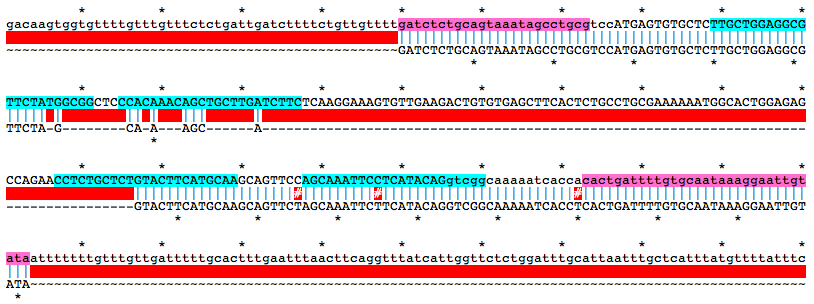
gatctctgcagtaaatagcctgcgtccATGAGTGTGCTCTTGCTGGAGGCGTTCTATGGCGGCTCCCACAAACAGCTGCTTGATCTTCTCAAGGAAAGTGTTGAAGACTGTGTGAGCTTCACTCTGCCTGCGAAAAAATGGCACTGGAGAGCCAGAACCTCTGCTCTGTACTTCATGCAAGCAGTTCCAGCAAATTCCTCATACAGgtcggcaaaaatcaccacactgattttgtgcaataaaggaattgtataMutation Area - Mutant DNA Sequence
GATCTCTGCAGTAAATAGCCTGCGTCCATGAGTGTGCTCTTGCTGGAGGCGTTCTAGCAAAGCAGTACTTCATGCAAGCAGTTCTAGCAAATTCTTCATACAGGTCGGCAAAAATCACCTCACTGATTTTGTGCAATAAAGGAATTGTATAGuide RNA Target Sites
TTGCTGGAGGCGTTCTATGGCGG
GAAGATCAAGCAGCTGTTTGTGG
TTGCATGAAGTACAGAGCAGAGG
AGCAAATTCCTCATACAGgtcgg
Genotyping Primers
f, gatctctgcagtaaatagcctgcg
r, tatacaattcctttattgcacaaaatcagtg
Alignment of WT and Mutant Sequences w/ gRNA Targets (cyan) and Genotyping Primers (pink) Shown on WT Sequence
Allele
103bpDWT Genotyping Size
254WT Protein Sequence
MSVLLLEAFYGGSHKQLLDLLKESVEDCVSFTLPAKKWHWRARTSALYFMQAVPANSSYR
VLFTSSVLNLAELVALRPDLGHLKKVLYFHENQLVYPVRKSQERDFQYGYNQILSCLVAD
VVVFNSSFNMESFLSSISTFMKTMPDHRPKDLERLIRPKCHVLHFPIRFPDVTRFLPAHK
RLRHPVRCDDIHAPAAKSHIQTSSPSSYPDVEPPEKMLNVAGTNQSHEPTSVTPHQETAS
PLCGHEGEKLRPLHIVWPHRWEHDKDPQLFFQTLLKLKDRQLSFEVSVLGETFTDVPDIF
SEAKEQLVDHIQHWGFMPSKEDYLKVLCQADVVVSTAKHEFFGVAMLEAVHCGCYPLCPK
ALVYPEIFPATYLYSTPEQLCKRLQEFCKRPQLARQHVVQVSLSSFSWDSLGDNFRSLLK
ADSGRMFTDKHMA-
Mutant Protein Sequence
MSVLLLEAF-Mutant Genotyping Size
151Behavior Data
Behavior Data Summary
NoneBehavior Data Description 1
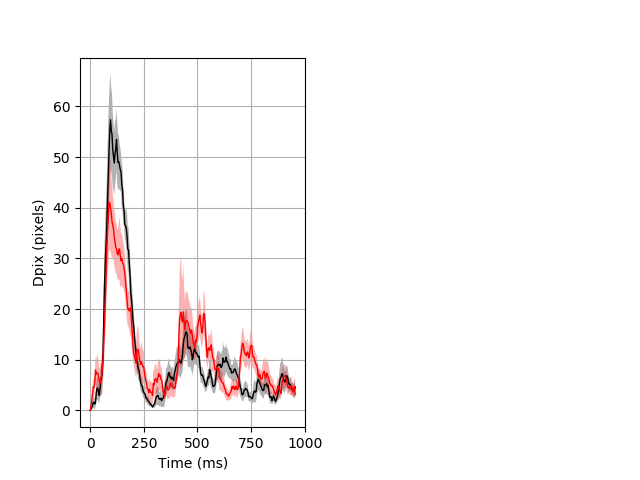
Dark flash block 1 start Merged section Pvalue = not significant 15 het vs 16 hom ribgraph_mean_ribbon_fullboutdata_dpix_a0darkflash103.pngBehavior Data Graph 1
Behavior Data Description 2
Day all prepulse tap Merged section Pvalue = not significant 15 het vs 16 hom ribgraph_mean_ribbon_fullboutdata_dpix_dayprepulseinhibition100d.pngBehavior Data Graph 2
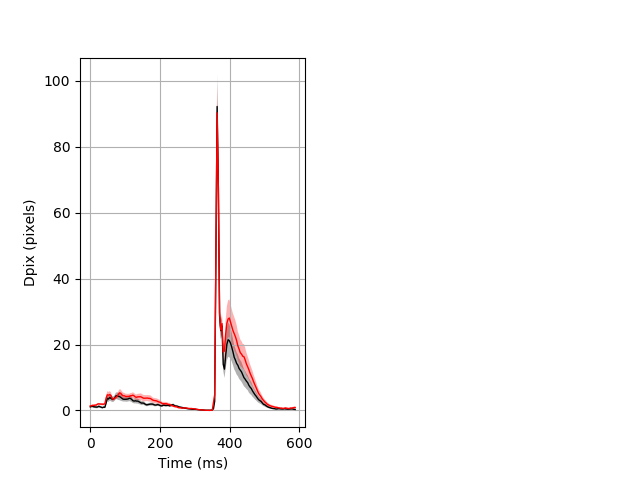
Behavior Data Description 3
Features of movement entire protocol Graph Pvalue = not significant Merged section Pvalue = not significant 15 het vs 16 hom ribgraph_mean_ribbonbout_aveboutdisp_10min_combo.pngBehavior Data Graph 3
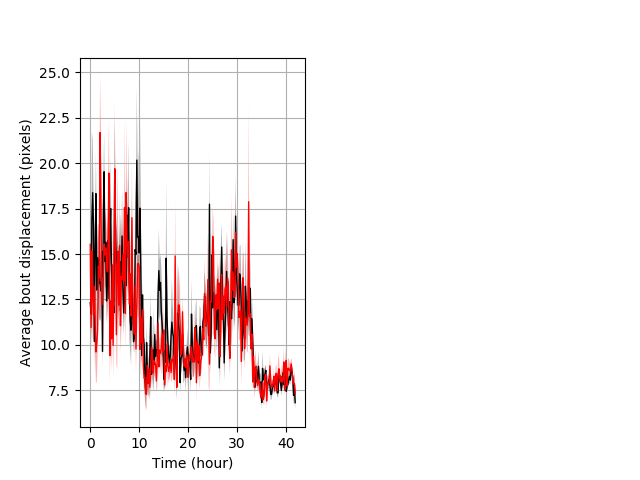
Behavior Data Description 4
Frequency of movement entire protocol Graph Pvalue = not significant Merged section Pvalue = not significant 15 het vs 16 hom ribgraph_mean_ribbonbout_dpixnumberofbouts_10min_combo.pngBehavior Data Graph 4
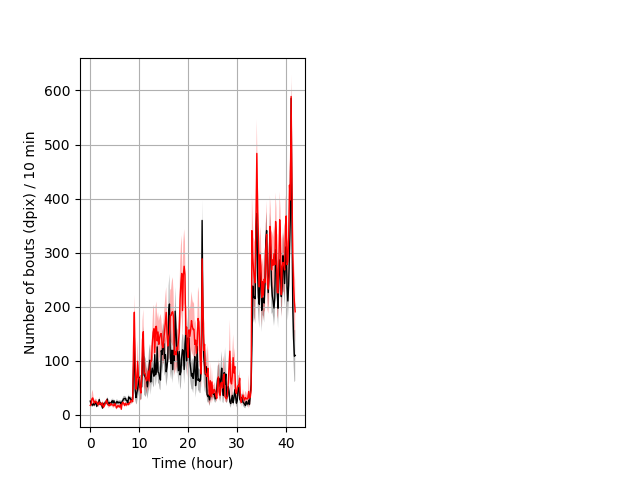
Behavior Data Description 5
Location in well entire protocol Graph Pvalue = not significant Merged section Pvalue = not significant 15 het vs 16 hom ribgraph_mean_ribbonbout_averhofrac_10min_combo.pngBehavior Data Graph 5
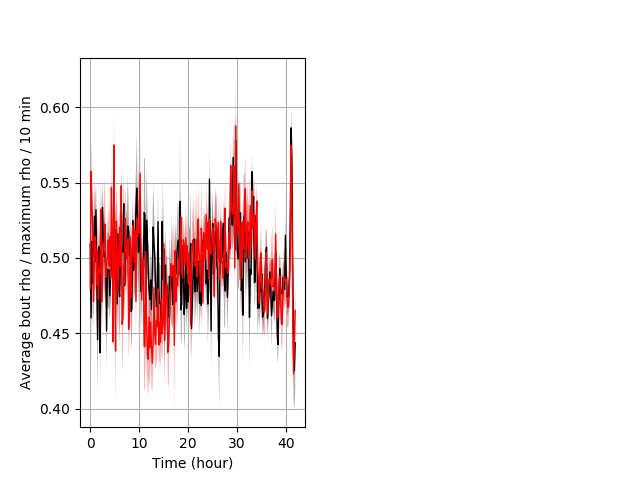
Behavior Data Description 6
NoneBehavior Data Graph 6
NoneBehavior Data Description 7
NoneBehavior Data Graph 7
NoneBehavior Data Description 8
NoneBehavior Data Graph 8
NoneBehavior Data Description 9
NoneBehavior Data Graph 9
NoneBehavior Data Description 10
NoneBehavior Data Graph 10
NoneBehavior Data Description 11
NoneBehavior Data Graph 11
NoneBehavior Data Description 12
NoneBehavior Data Graph 12
NoneBehavior Data Description 13
NoneBehavior Data Graph 13
NoneBehavior Data Description 14
NoneBehavior Data Graph 14
None