Locus Rank
33Gene
Id
262Gene Name
CSMD3Duplicated
FalseMaternal
FalseGene Not Within Locus (Nearby
TrueGene Description
Mouse Phenotype
Additional Image
Allen Brain
Allen Regions
Zfin In Situ
Zfin Brain Areas
Published Zebrafish Pehnotype
Not In Allen Brain
FalseZFIN Link
NoneAllen Link
http://mouse.brain-map.org/gene/show/88573More Additional Images
Papers
Locus Rank
Locus Rank
33Genes in Region
No gene in locus itself, KCNV1, CSMD3, and SYBU are far away on each side of the gene desert.Associated Snps
GWAS Region (hg19)
chr8:111460061-111630761Genes Skipping
Ricopili Plot Surrounding Area
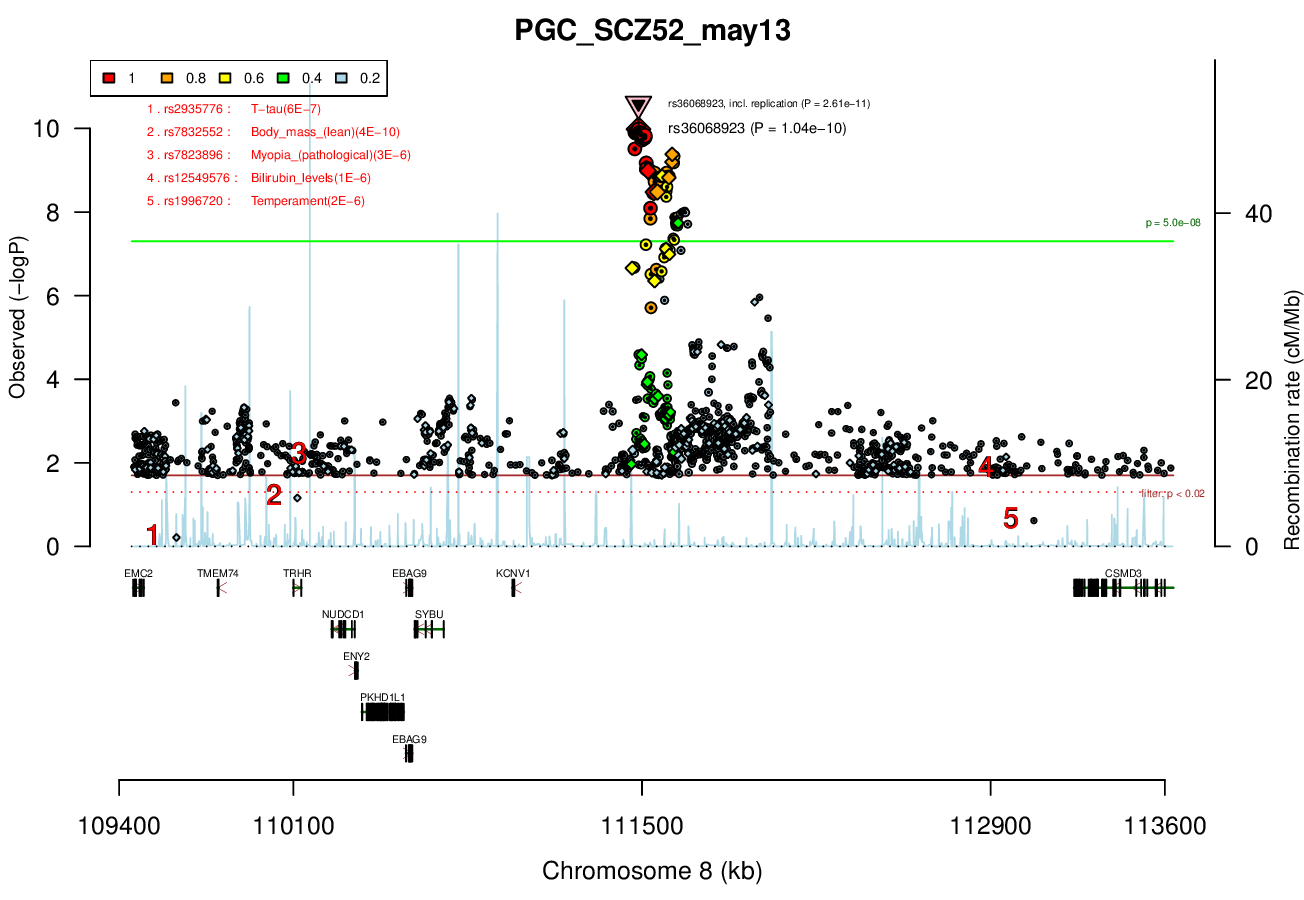
Distance On Each Side Of Locus In Above Plot
2 MBRicopili Plot
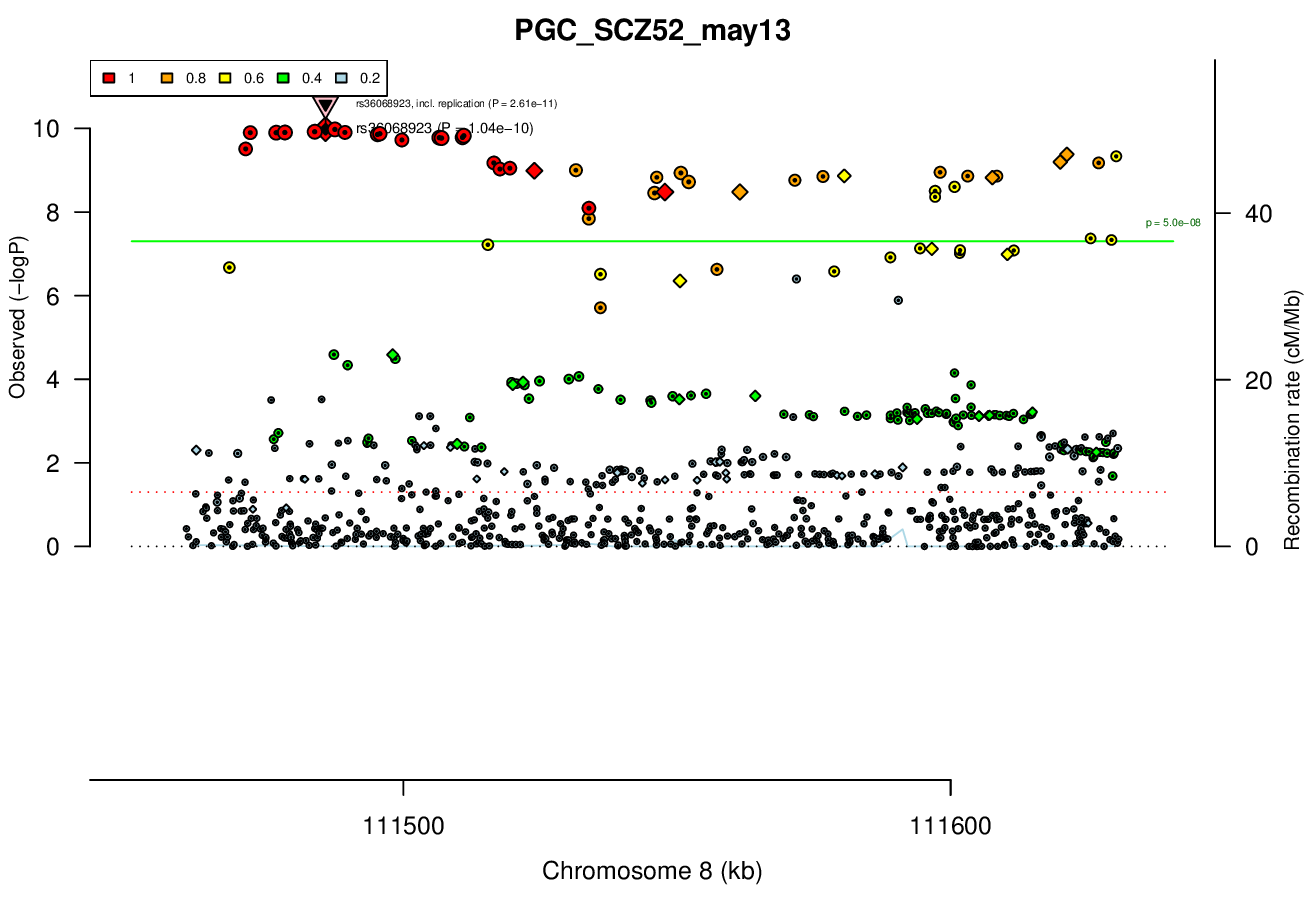
Gwas And Surrounding Region Pic
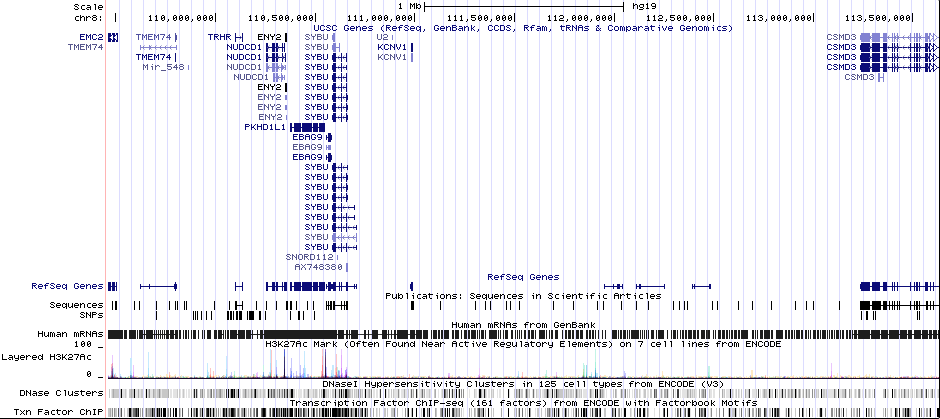
Gwas Region Pic
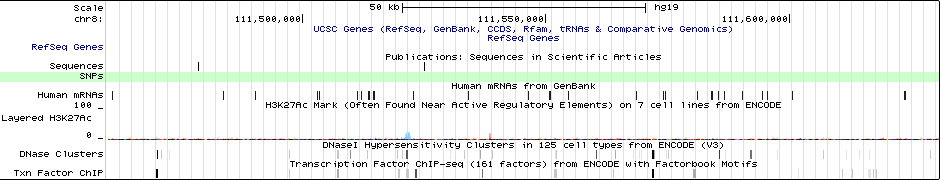
Sequence 1
Ensembl Gene Name
csmd3Harvard Allele
a254Mutation Area - WT DNA Sequence
accatgaatgtagtcttaaaagtgttccttttttctgccttttcAGAATTGCAGAGCAGTTCTTGTGGGAACCCTGGCGTCCCGCCTAAAGGCATTCTGTACGGGACGCGCTTTAATGTAGGCGACAAGATCCGCTACAGCTGTGTGACCGGCTACGTGCTGGACGGCCATCCGCAGCTCACCTGTGTGACTAATGCAGGCAACGCTGCCGTATGGGACTTTCCAGTGCCCATCTGTAGAGgtaagcatgcaactgtgcgtcgtgcatgatgaagacactgtcagcgMutation Area - Mutant DNA Sequence
ACCATGAATGTAGTCTTAAAAGTGTTCCTTTTTTCTGCCTTTTCAGAATTGCAGAGCAGTTCTTGTGGGAACCCTGGCGTCCCGCCTAAAGGCATTCTGTACGGGACGCGCTTTAATGTAGGCGACAAGATCCGCTACAGCTGTGTGACCGGCTACGTGCTGGACGGCCATCCATCTGTAGAGGTAAGCATGCAACTGTGCGTCGTGCATGATGAAGACACTGTCAGCGGuide RNA Target Sites
GAATTGCAGAGCAGTTCTTGTGG
AGCCGGTCACACAGCTGTAGCGG
TTAGTCACACAGGTGAGCTGCGG
TTTCCAGTGCCCATCTGTAGAGg
Genotyping Primers
f, accatgaatgtagtcttaaaagtgttcc
r, cgctgacagtgtcttcatcatgc
Alignment of WT and Mutant Sequences w/ gRNA Targets (cyan) and Genotyping Primers (pink) Shown on WT Sequence
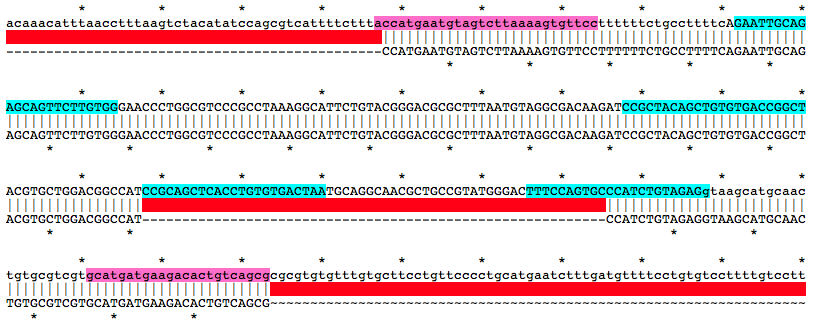
Allele
58bpDWT Genotyping Size
287WT Protein Sequence
XELQSSSCGNPGVPPKGILYGTRFNVGDKIRYSCVTGYVLDGHPQLTCVTNAGNAAVWDF
PVPICRAEDTCGDTLRGSSGIITSPNFPSEYYNSADCTWTILADPGDTISIIFTDFQTEE
KYDYLEVEGSEPPTIWLSGMNVPSPIVSNKNWLRLHFVTDSNHRYKGFSAHYQVKRSVDF
KSRGVKLFPGKDNSNKFSILNEGGIKQASNSCPDPGEPENGKRIGNDFSIGATAEFTCDE
DHVLQGSKSITCQRVAEVFAAWTDHRPVCRVKTCGSNLQGPSGTFSSPNFPIQYESNSQC
VWIITASNLNKVIQINFEDFDLELGYDTLTIGDGGEVGDPKTILQVLTGSFVPDLIVSMT
HQMWLHLQSDESVGSIGFKINYKEIDKESCGDPGTPLYGMREGDGFSNGDMLRFECQFGF
ELIGEKTISCQNNNQWSANIPICIFPCLSNFTAPMGTVLSPDYPEGYGNNLNCVWLILSE
PGSRIHLAFNDFDLEAPYDILTVKDGEMLDSSVLGRFTGAEVPSHLTSNSNILRLEFQAD
HSMSGRGFNITYTTFGHNECPDPGIPLNARRFGDSFQLGSSISIICEDGFIKTQGAETIT
CELNGGKVMWSGPIPKCEAPCGGHFSAPSGVILSPGWPGYYKDSLNCEWVIEAEPGRSIK
ITFEKFQTELNCDFLELHDGPNLLSPLIGSFNGTQVPQFLFSSGNHLYMLFTTDNSRSNS
GFKILYESVTVDAYSCLDPGIPVNGLRYGQDFSIGSTVSFGCDSGYRLSHEEPLVCEKNH
WWSHPLPTCDALCGGDVRGPGGTILSPGYPEVYPSSLNCTWTVEVSHGKGVQFTFNTFHL
EDHHDYLLITENGSFVQPLARLTGAELPLPINAGLYGNFKAQLRFISDFSISYEGFNITF
SEYNLEPCEDPGVPHYGRRNGYSFGIGDTLTFSCNLGYRLEGAPEIICLGGGRRMWSAPL
PRCVAECGSSVINNEGILLSPNYPMNYENNHECIYSIQVQAGKGINISARTFHLAQGDIL
KIYDGKDNAAHMLGAFTGSSMLGLTLISTSNHLWLEFYSDAEATGEGFKLVYSSFELSHC
EDPGVPQFGFKVNDQGHFSGSTITYKCDPGYTLHGSGVLKCMTGERRAWDNALPSCIAEC
GGRFKGETSGRILSPGYPFPYDNNLRCTWMVEVDSGNIISLQFLAFDTEASHDILRVWDG
PPENEMSLREVSGSLLPENVHSTLNMVSIQFETDFYISKSGFAIEFSSSVATACRDPGVP
MNGSRNGDGREPGDTVTFQCDPGYELQGDVKITCIQVENRYYWQPSPPVCIAPCGGNLTG
SSGFILSPNFPHPYPHSKDCDWLIAVNSDYVLSLAFISFSIEPNYDFLYIYDGADSNSPL
IGSFQDSKLPERIESSSNTMYLAFRSDGSVSYTGFHLEYKAKLRESCFDPGNVMNGTRLG
SDYKLGSTVTFYCEAGYVLQGYSTLTCIMGDDGRPGWNRGLPSCQAPCGSRSTGTDGTVL
SPNFPRNYTTGQSCVYTIAVPREFVLFGQFVLFQTSLNDVVDIYDGPTQQSSLLSSLSGS
HSGESLPLSTGNQITIKFSTVGPETAKGFHFVYQAVPRTSATQCSSVPEPRFGKRIGNNF
AVGASVMFECNPGYTLHGSTAIHCETVPDALAQWNDTLPTCIVPCGGILTMRKGTILSPG
YPEPYDNNLNCVWKVSVPEGAGIQIQVVSFATEHNWDSLDFYDGADNNAPRLGSYSGTTI
PQLLNSTSNNLYLNFVSDISVSAAGFHLEYTAIGLDTCPEPQTPTYGIKIGDRYMVGDVV
MFQCEQGYSLQGNAHITCMPGPVRRWNYPVPLCLAQCGGAMTEVSGVILSPGFPGNYPSG
LDCTWTITLPIGFGIHVQFLNFSTEAIHDYLEIRSGPSETGTVIDRFSGPQVPESLFSTT
HETSFFFHSDYSQNKPGFHITYQAYQLQSCPDPRPFRNGIVIGSDFSVGMTVSFECLPGY
SLIGETSLTCLHGISRNWNNPIPRCEALCGGNITSMNGTIFSPGHPAEYPNFQDCVWSVR
VPPGNGIYINFTVLSTEPIYDYITVWDGPDQSSPQIGQFSGNTALESVYSTSNQILIKFH
SDFSGSGFFVLSYHAYQLRVCQPPLEVANADILMEDNELEIGDIIRYRCHPGFTLVGSEI
LTCRLGERLQMDGHPPSCQVQCPAHDVRFDSSGVILSPGYPDSYPNLQMCSWSINVEKGY
NITLFFEFFQTEKEYDILEVFDGPTIHSQTLATLSGDLPTPFNMTTSGHQLLLRWSADHG
TNKKGFSIRYVALYCSTPDSPLHGSISSQTGGHLNSLVRWTCDRGYRLIGKSTAVCKKTP
FGFYDWDAPVPACQAVSCGVPKAPVNGGVLAMDYSVGTRVTYFCSDGYRLSSKELTSAVC
QPDGTWSNHNKVPRCSVVVCPSIGSFSLEHGRWRIVNGSHYEYRTKIIFTCDPGYYRLGP
AYIQCMANGAWSWKNERPHCQIISCGDLATPPSGKKIGTQTSFGATAIFTCDNGYMLVGS
SVRECLSSGLWSGTETRCLAGHCGLPEQIVNGQVIGENFGYRDTVVYQCVSGFRLIGSSV
RICQQDHNWSGQLPICVPISCGHPGSPIYGRTIGNGFNLNDVVSFSCNMGYVMEGPSRAQ
CQANRQWSQPPPTCKVVNCSDPGIPANSIRQSKIEHGNFTFGTVVFYDCNPGYYLFGSPV
LTCQPTGQWDKPLPECIVVDCGHPGSPPNGLLSGDKFTFGATVRYSCTGGRQLKGESSRT
CQLNGHWSAPMPFCSGDSSGTCGDPGTPSHGSRDESDFRIRSKVHFSCSVGYELFGSVER
MCFPNGTWSGTQPSCKPVQCGNPGTPSNGRVYRLDGTTFSHSVIYSCMDGYLLSGSTTRQ
CQANGTWSGTAPNCTLINCGEPGVPANGLRYGEDFTIAQNITFMCQPGYTMEAESFSTIT
CTGNGTWNAPVPFCKAVTCTAPPPISNGLLEGRDFEWGTSVSYSCSPGYELSFPAILTCV
GNGTWSGEVPQCLPKFCGDPGTPAFGSREGRSFIYQSEVSFSCMAPYIPVGSTTRICQTD
GTWSGSQPRCIEPTRTTCENPGTPRYGSLNRTFGFKVGSMVAFQCQPGHLLQGSTTRLCQ
ADLTWSGTQPECIPHSCKQPESPPHVDVVGMDLPSYGYTLIYSCQSGFFLSGGSEHRVCR
SDGTWTGKMPVCRGSEHIPLCFKKRSVIQLKLCTHEATIAGSKLSEKPIKPVPGTPSPKL
NVPDDVFAPNYIWKGSYNYKGRKQPMTLSITSFNTTTGRVNVTLTSSNMELLLSGVYKSQ
EARLMLLVYHVKASSSSTLSKIKEEHWTLDGFVSAEPDGSTYVFQGFIQGKDYGQFGLQR
LGLNMSESNNSSNPPHGTNSSSVAIAILVPFFALIFAGFGFYLYKQRTTPKTQYTGCSVH
ENNNGQAAFENPMYNTKPEVKNVRFDPNLNTVCTMV
Mutant Protein Sequence
XELQSSSCGNPGVPPKGILYGTRFNVGDKIRYSCVTGYVLDGHPSVELRTHVVIL-In Situ Probe Sequence
GCGGCACCATCTCCAGTCCAGACTTTCAGAAGGACTATCAGAGCAGTGGTGAATGTACGTGGACCATCCTGGCTGATCCAGGAGACACCATCTCTCTGGTCTTCACTGAGTTCCAGATGGACAGCAAACTGGACTCTTTAGAAGTGGAGGGATCAGATCCACCTACCATATGGTTATCAGGAATTAATATTCCTGTGCCAATAGTGAGCAACAAGAACTGGCTACGGCTTCATTTCGTAACAGACAACAGCCATCGGCATAAAGGATTCAGCGCTCAATATCAAGCAGTGAAACGATCTGGGGATTTTAAGTCAAGAGGAGTCAAATTATTACCTGGAAAAGACAACACAAGCAAGTTTTCCATCATGAATGAAGGGATGGTCAAGCAGGTCTCCAACTTCTGCCCAGATCCTGGAGAACCAGAGAACGGCAAACGCATTGGCACTGATTACAGCATCGGAGCGACGGTTCAGTTCACCTGTGACGGAGATCATGTGCTTCAGGGATCCAAAAGCATCACGTGTCAGAGAGTGGCGGAGGTGTTTGCAGCCTGGAGCGACCACAGACCCGTCTGCAAAGTGAAAACATGTGGGATGAATCTCTTGGGTCCTTCAGGAACGTTTACGTCTCCTAATTTCCCCATCCAGTATGAAAGCAACTCTCAGTGTGTTTGGATCATCACCGCCAGTAACCCCAATAAGGTTATTCAAATCAATTTCGAAGAGTTCGACTTGGAACTCGGATATGATACATTGACTGTCGGAGACGGCGGAGAAGTTGGAGMutant Genotyping Size
229Behavior Data
Behavior Data Summary
NoneBehavior Data Description 1
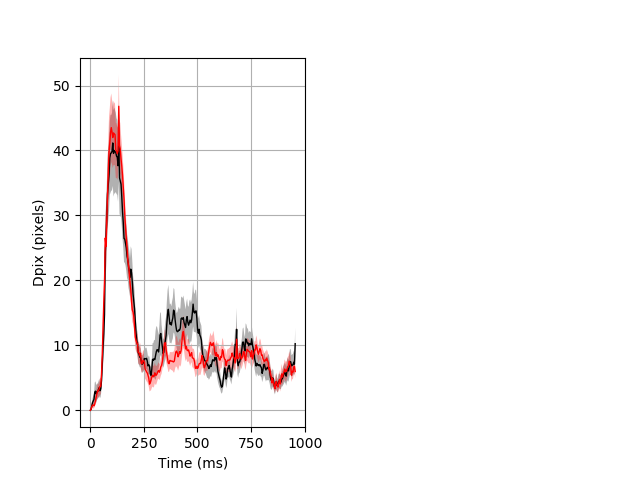
Dark flash block 1 start Merged section Pvalue = 0.0268 30 het vs 29 hom ribgraph_mean_ribbon_fullboutdata_dpix_a0darkflash103.pngBehavior Data Graph 1
Behavior Data Description 2
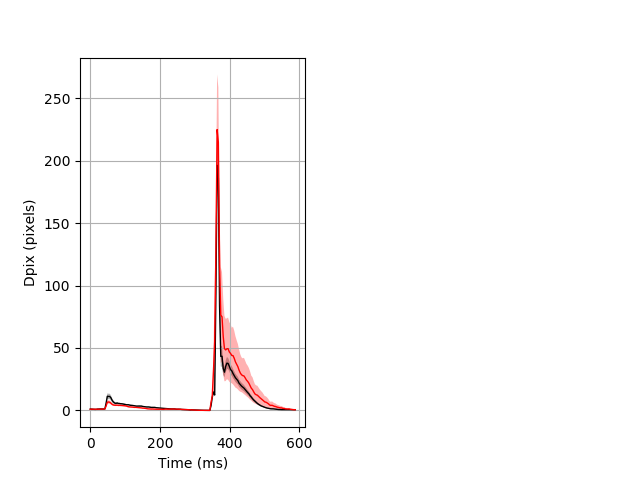
Day all prepulse tap Merged section Pvalue = 0.0129 30 het vs 29 hom ribgraph_mean_ribbon_fullboutdata_dpix_dayprepulseinhibition100d.pngBehavior Data Graph 2
Behavior Data Description 3
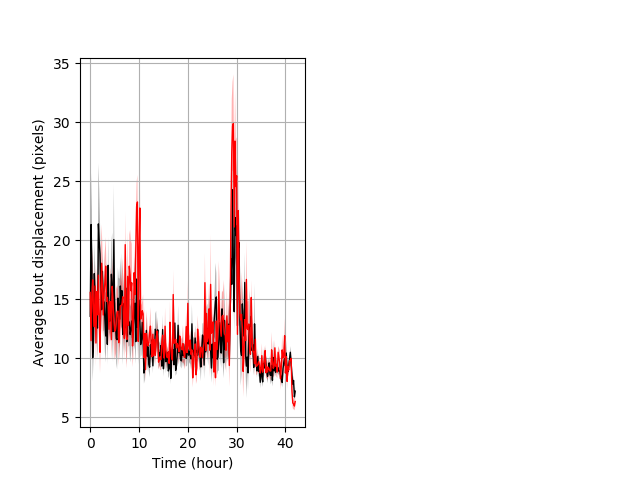
Features of movement entire protocol Graph Pvalue = not significant Merged section Pvalue = not significant 18 het vs 23 hom ribgraph_mean_ribbonbout_aveboutdisp_10min_combo.pngBehavior Data Graph 3
Behavior Data Description 4
Frequency of movement entire protocol Graph Pvalue = 0.0188 Merged section Pvalue = 0.00652 30 het vs 29 hom ribgraph_mean_ribbonbout_dpixnumberofbouts_10min_combo.pngBehavior Data Graph 4
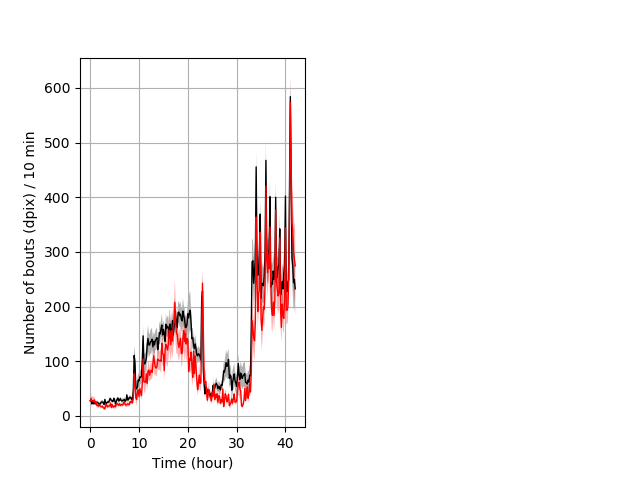
Behavior Data Description 5
Location in well entire protocol Graph Pvalue = not significant Merged section Pvalue = 0.033 30 het vs 29 hom ribgraph_mean_ribbonbout_averhofrac_10min_combo.pngBehavior Data Graph 5
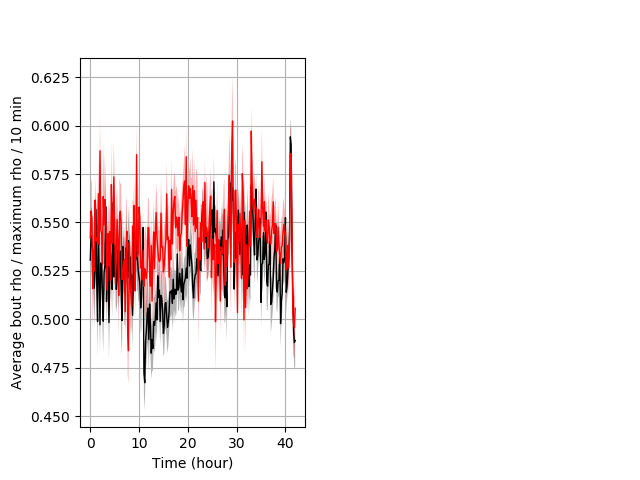
Behavior Data Description 6
Features of movement Graph Pvalue = 0.001 18 het vs 23 hom ribgraph_mean_ribbonbout_aveboutvel_min_day1mornstim.pngBehavior Data Graph 6
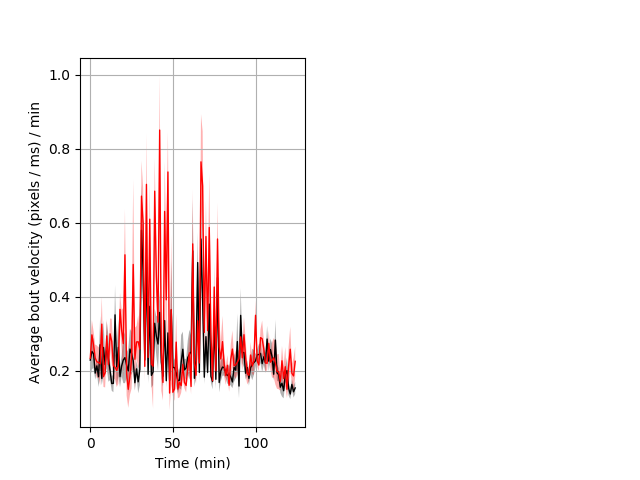
Behavior Data Description 7
Frequency of movement Graph Pvalue = 9.45e-05 30 het vs 29 hom ribgraph_mean_ribbon_distminper_10min_combo.pngBehavior Data Graph 7
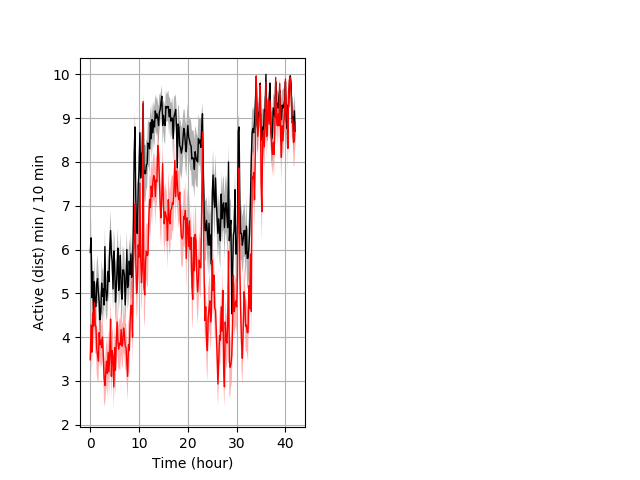
Behavior Data Description 8
Weak tap Graph Pvalue = 0.0116 18 het vs 23 hom boxgraph_pd__ribgraph_mean_ribbon_peakspeed_nightprepulseinhibition101.pngBehavior Data Graph 8
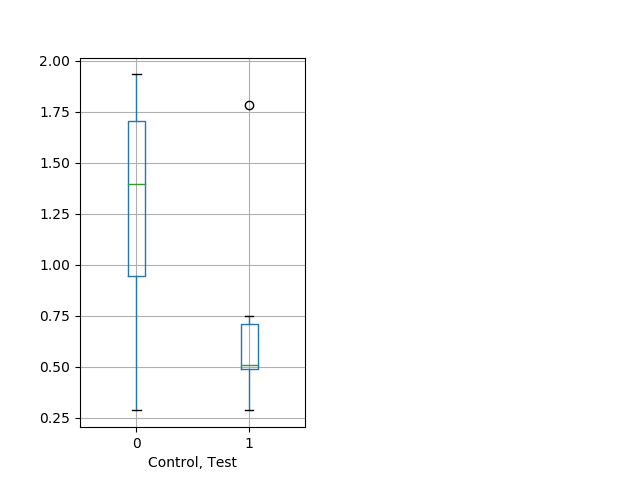
Behavior Data Description 9
NoneBehavior Data Graph 9
NoneBehavior Data Description 10
NoneBehavior Data Graph 10
NoneBehavior Data Description 11
NoneBehavior Data Graph 11
NoneBehavior Data Description 12
NoneBehavior Data Graph 12
NoneBehavior Data Description 13
NoneBehavior Data Graph 13
NoneBehavior Data Description 14
NoneBehavior Data Graph 14
None