Locus Rank
45Gene
Id
190Gene Name
RORADuplicated
TrueMaternal
FalseGene Not Within Locus (Nearby
TrueGene Description
Mouse Phenotype
Additional Image
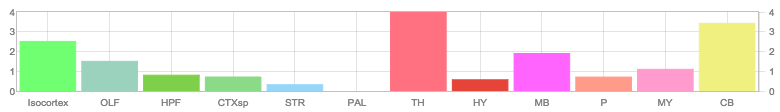
Allen Brain
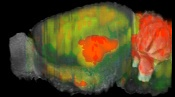
Allen Regions
Zfin In Situ
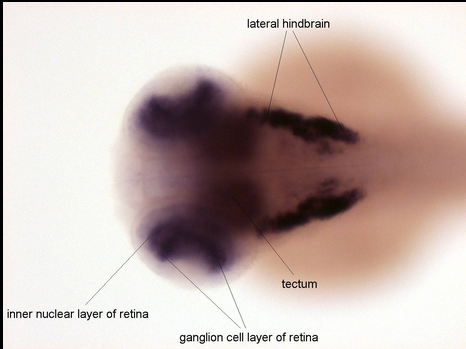
Zfin Brain Areas
Published Zebrafish Pehnotype
Not In Allen Brain
FalseZFIN Link
http://zfin.org/search?q=rora&fq=category%3A%22Gene+%2F+Transcript%22&category=Gene+%2F+Transcript&fq=type%3A%22Gene%22Allen Link
http://mouse.brain-map.org/gene/show/19646More Additional Images
Papers
http://www.ncbi.nlm.nih.gov/pubmed/24500708Locus Rank
Locus Rank
45Genes in Region
RORA, VPC13CAssociated Snps
rs12903146GWAS Region (hg19)
chr15:61831663-61909663Genes Skipping
VPS13C was made, but then the zebrafish genome changed and a new/dominant isoform was predicted that was not eliminated in the tested allele. Maybe worth remaking, although RORA has additional associations with schizophrenia in other studies, as well as associations with autism and depression.Ricopili Plot Surrounding Area
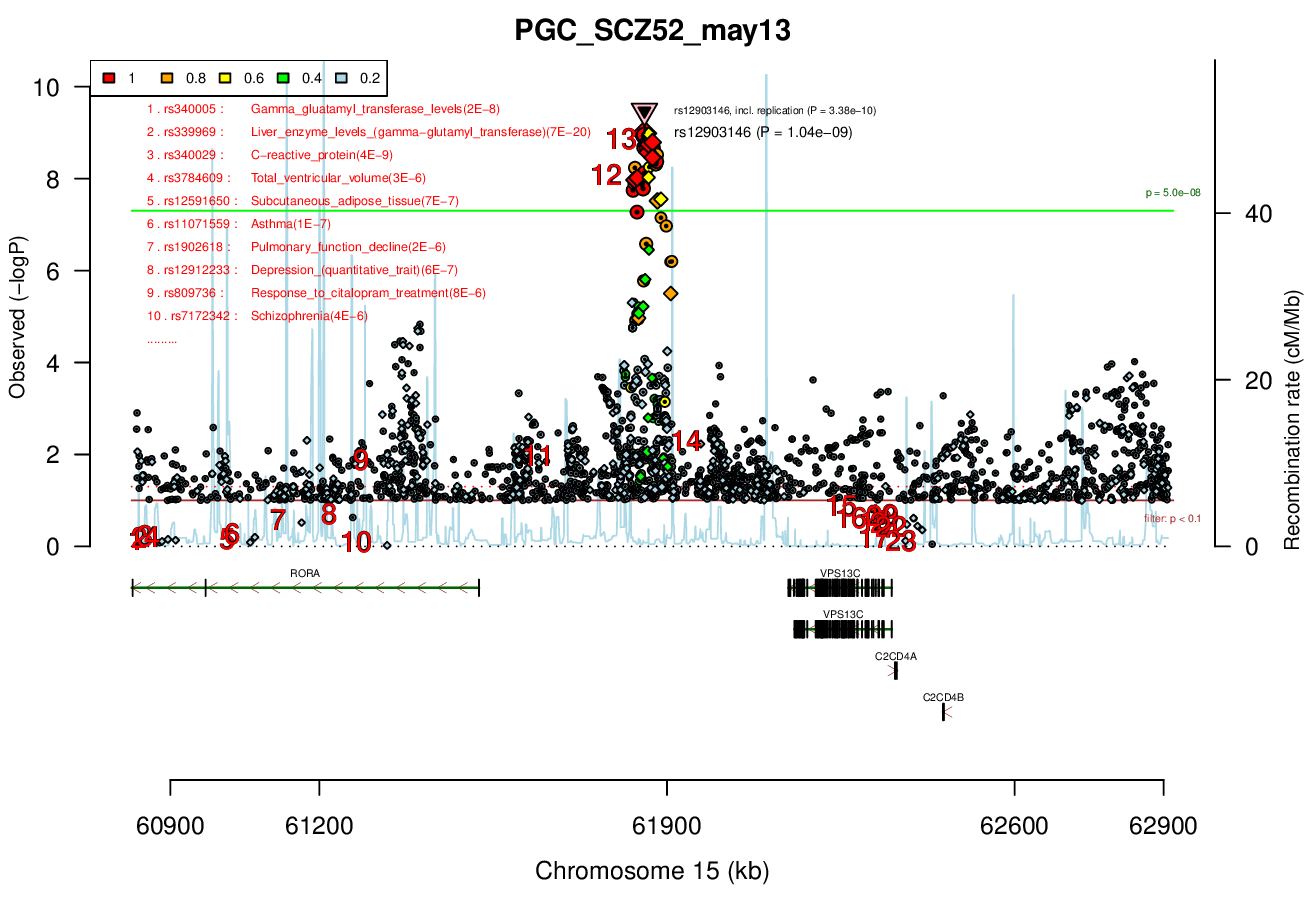
Distance On Each Side Of Locus In Above Plot
1 MBRicopili Plot
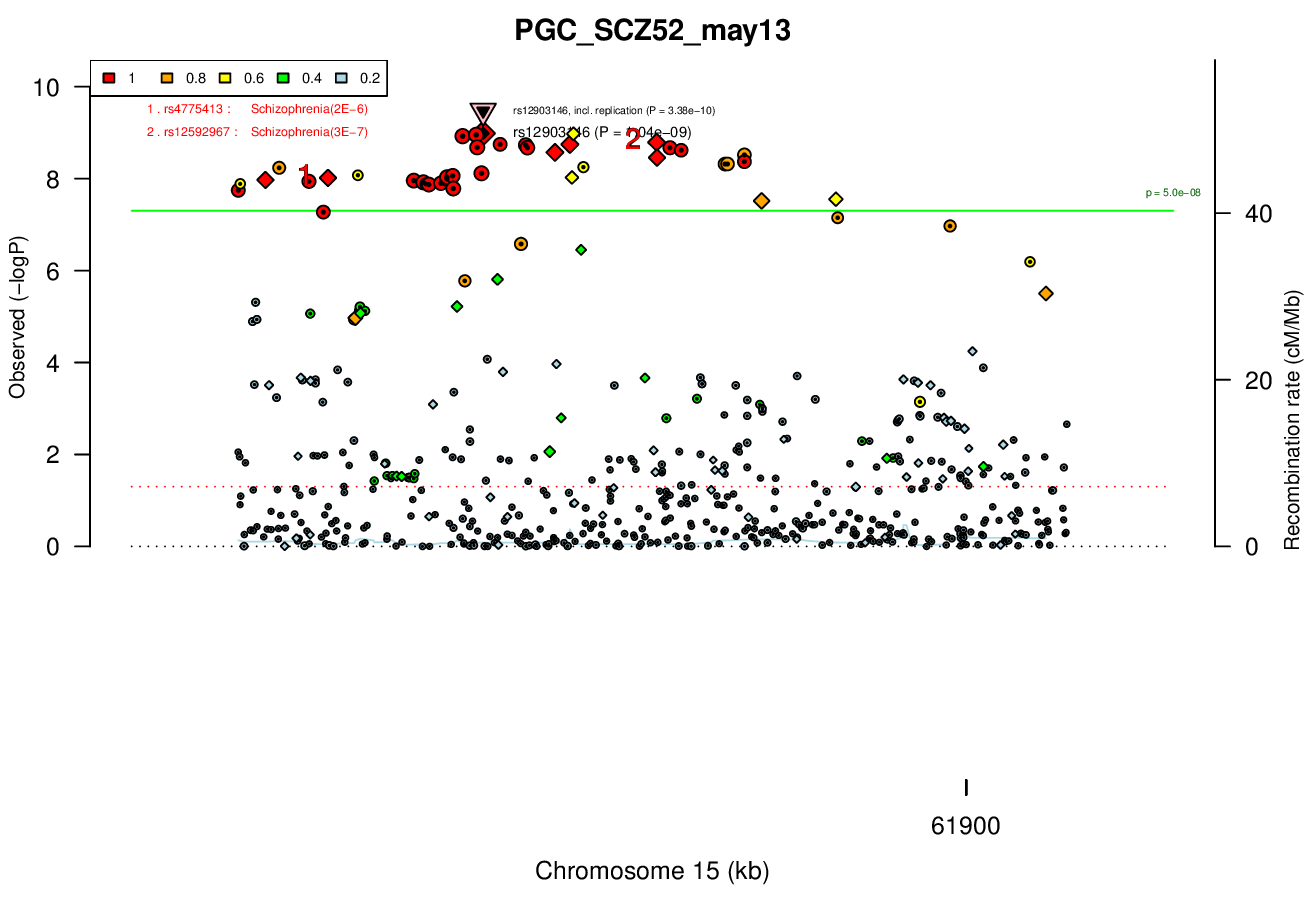
Gwas And Surrounding Region Pic
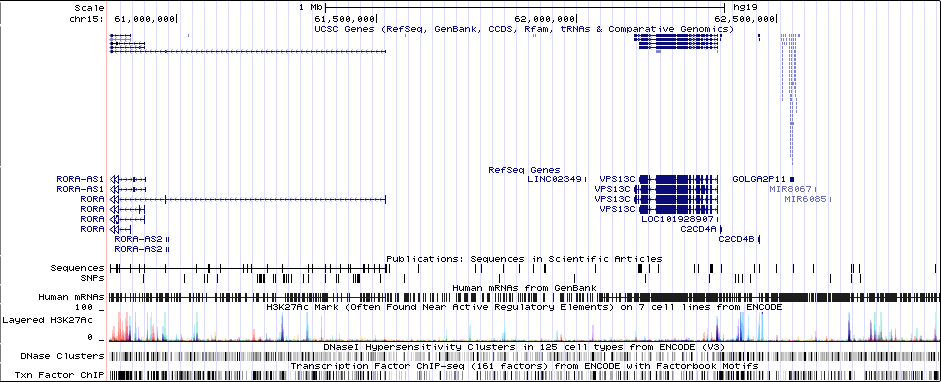
Gwas Region Pic
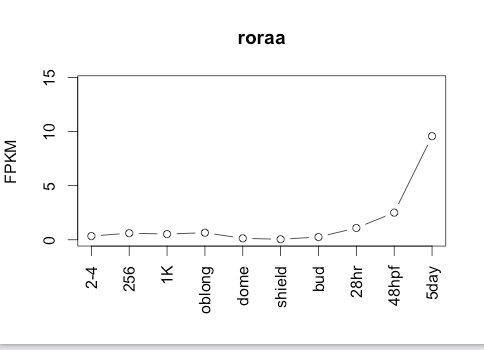
Sequence 1
Ensembl Gene Name
roraaHarvard Allele
a319Mutation Area - WT DNA Sequence
AGCGGGACCACCAGCAGCAGCCCGGTGAAGCCGAACCCCTTACCCCAACTTATGGCCTCTCCACCAATGGTCTGACCGAGCTCCACGATGATCTGAGTGGATACATGAATGGCCACACGCCGGATGGCACCAAGCCTGACTCAGGAGTCAGCAGCTTCTACCTGGACATCCAACCCTCACCTGATCAATCCGGCCTGGACATCAATGGCATCAAGCCAGAGCCCATCTGCGACTTTACCCCTGGCTCAGGCTTCTTCCCCTACTGCTCCTTCACCAATGGGGAGACGTCCCCCACCGTTTCCATGGCTGAGTTGGgtaagtgatcattcaaatgaatggtaaagtggMutation Area - Mutant DNA Sequence
AGCGGGACCACCAGCAGCAGCCCGGTGAAGCCGAACCCCTTACCCCAATGGGGAGACGTCCCCCACCGTTTCCATGGCTGAGTTGGgtaagtgatcattcaaatgaatggtaaagtggGuide RNA Target Sites
GGTGGAGAGGCCATAAGTTGGGG
GAGCTCCACGATGATCTGAGTGG
CATTGGTGAAGGAGCAGTAGGGG
ACTCAGCCATGGAAACGGTGGGG
Genotyping Primers
f, AGCGGGACCACCAGCAGCAG
r, ccactttaccattcatttgaatgatcac
wt_f, TGGATACATGAATGGCCACACGCCG
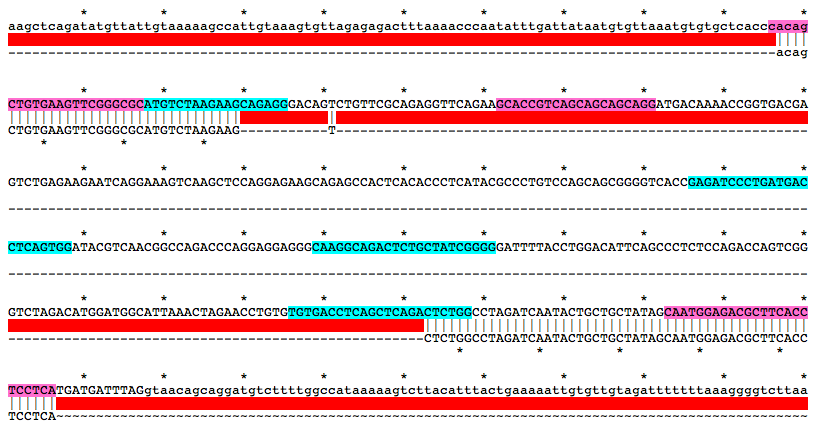
Alignment of WT and Mutant Sequences w/ gRNA Targets (cyan) and Genotyping Primers (pink) Shown on WT Sequence

Allele
229bpDWT Genotyping Size
250WT Protein Sequence
MSESHGFWCGHASAAERQSYAGQLLLKRQWTCTALALAAPSAYGPPSLMMYFVISAMKAQ
IEIIPCKICGDKSSGIHYGVITCEGCKGFFRRSQQSNAAYSCPRQKNCLIDRTSRNRCQH
CRLQKCLAVGMSRDAVKFGRMSKKQRDSLYAEVQKHRLQQQQRDHQQQPGEAEPLTPTYG
LSTNGLTELHDDLSGYMNGHTPDGTKPDSGVSSFYLDIQPSPDQSGLDINGIKPEPICDF
TPGSGFFPYCSFTNGETSPTVSMAELEHLAQNISKSHMETCQYLREELQQMTWQAFLQEE
VENYQSKPREVMWQLCAIKITEAIQYVVEFAKRIDGFMELCQNDQIVLLKAGSLEVVFVR
MCRAFDPQNNTVYFDGKYAGPDVFKSLGCDDLISSVFEFGKNLCSMHLSEDEIALFSAFV
LMSADRSWLQEKVKVEKLQQKIQLALQHVLQKNHREDGILTKLICKVSTLRALCSRHTEK
LTAFKAIYPDIVRAHFPPLYKELFGSDFEQSMPVDG-
Mutant Protein Sequence
MSESHGFWCGHASAAERQSYAGQLLLKRQWTCTALALAAPSAYGPPSLMMYFVISAMKAQ
IEIIPCKICGDKSSGIHYGVITCEGCKGFFRRSQQSNAAYSCPRQKNCLIDRTSRNRCQH
CRLQKCLAVGMSRDAVKFGRMSKKQRDSLYAEVQKHRLQQQQRDHQQQPGEAEPLTPM
GRRPPPFPWLSWNIWPRTFPSPTWRRVST-
In Situ Probe Sequence
GGCACCAAGCCTGACTCAGGAGTCAGCAGCTTCTACCTGGACATCCAACCCTCACCTGATCAATCCGGCCTGGACATCAATGGCATCAAGCCAGAGCCCATCTGCGACTTTACCCCTGGCTCAGGCTTCTTCCCCTACTGCTCCTTCACCAATGGGGAGACGTCCCCCACCGTTTCCATGGCTGAGTTGGAACATCTGGCCCAGAACATTTCCAAGTCCCACATGGAGACGTGTCAGTACCTGAGAGAAGAACTTCAGCAGATGACCTGGCAGGCGTTCCTTCAGGAGGAGGTGGAGAACTACCAGAGCAAGCCCAGGGAAGTCATGTGGCAGTTGTGTGCCATCAAAATCACAGAGGCCATTCAGTATGTGGTGGAGTTTGCCAAGCGCATCGACGGCTTCATGGAGCTCTGTCAGAATGATCAGATAGTGCTTCTCAAAGCAGGCTCTTTGGAGGTGGTGTTCGTCAGAATGTGCCGTGCCTTCGACCCTCAGAACAACACAGTCTATTTCGATGGAAAGTATGCCGGGCCTGATGTCTTTAAATCATTAGGCTGCGACGACTTGATCAGCTCTGTCTTCGAGTTTGGGAAAAACTTGTGTTCTATGCACCTCTCTGAAGACGAGATCGCCCTGTTCTCCGCCTTCGTCTTGATGTCTGCCGACCGATCCTGGCTCCAGGAGAAGGTGAAGGTGGAGAAACTCCAGCAGAAGATCCAGCTGGCCCTCCAACATGTTCTGCAGAAAAACCACAGAGAGGACGGCATACTGACAAAGTTGATATGCAAAGTGTCCACGCTGCGAGCCCTGTGCAGCCGGCATACGGAAAAGCTGACGGCGTTCAAAGCAATATACCCAGACATTGTCCGCGCACATTTCCCCCCTTTGTACAAGGAGTTATTCGGATCGGACTTTGAGCAGTCGATGCCCGMutant Genotyping Size
118Ribosome Profiling Development
Transcript Plot
Sequence 2
Ensembl Gene Name
rorabHarvard Allele
a320Mutation Area - WT DNA Sequence
cacagCTGTGAAGTTCGGGCGCATGTCTAAGAAGCAGAGGGACAGTCTGTTCGCAGAGGTTCAGAAGCACCGTCAGCAGCAGCAGGATGACAAAACCGGTGACGAGTCTGAGAAGAATCAGGAAAGTCAAGCTCCAGGAGAAGCAGAGCCACTCACACCCTCATACGCCCTGTCCAGCAGCGGGGTCACCGAGATCCCTGATGACCTCAGTGGATACGTCAACGGCCAGACCCAGGAGGAGGGCAAGGCAGACTCTGCTATCGGGGGATTTTACCTGGACATTCAGCCCTCTCCAGACCAGTCGGGTCTAGACATGGATGGCATTAAACTAGAACCTGTGTGTGACCTCAGCTCAGACTCTGGCCTAGATCAATACTGCTGCTATAGCAATGGAGACGCTTCACCTCCTCAMutation Area - Mutant DNA Sequence
cacagCTGTGAAGTTCGGGCGCATGTCTAAGAAGTCTCTGGCCTAGATCAATACTGCTGCTATAGCAATGGAGACGCTTCACCTCCTCAGuide RNA Target Sites
GGCGCATGTCTAAGAAGCAGAGG
GAGATCCCTGATGACCTCAGTGG
CAAGGCAGACTCTGCTATCGGGG
TGTGACCTCAGCTCAGACTCTGG
Genotyping Primers
f, cacagCTGTGAAGTTCGGGCGC
r, TGAGGAGGTGAAGCGTCTCCATTG
wt_f, GCACCGTCAGCAGCAGCAGG
These three primers cannot be mixed together. A separate PCR must be run with wt_f and r.
Alignment of WT and Mutant Sequences w/ gRNA Targets (cyan) and Genotyping Primers (pink) Shown on WT Sequence
Allele
322bpDWT Genotyping Size
345WT Protein Sequence
MYLMITAMKAQIESIPCKICGDKSSGIHYGVITCEGCKGFFRRSQQGTVSYSCPRQKSCL
IDRTSRNRCQHCRLQKCLAVGMSRDAVKFGRMSKKQRDSLFAEVQKHRQQQQDDKTGDES
EKNQESQAPGEAEPLTPSYALSSSGVTEIPDDLSGYVNGQTQEEGKADSAIGGFYLDIQP
SPDQSGLDMDGIKLEPVCDLSSDSGLDQYCCYSNGDASPPHDDLEHLSENICKSHLETCQ
YLREELQPSNWQTVLQSNLEAYQKKSQEDMWQLCAVKVTEAVQYVVEFAKRIDGFMELCQ
NDQIVLLKAGSLEVVFVRMCRAYNSQNNTVFFDSKYAGPEVFKALGCDDLISSVFEFAKS
LNSLQLSEDEIGLFSAYVLMSADRSWLQEKTRVEKLQQKIKIALQNLLQKNKRDEGILTK
LVCKVSTVRMLCRRHMEKLSAFRALYPEMVHTRFPPLYKELFGSDFEQILPQFMYTAFSQ
SSPATLCPNGTV-
Mutant Protein Sequence
MYLMITAMKAQIESIPCKICGDKSSGIHYGVITCEGCKGFFRRSQQGTVSYSCPRQKSCL
IDRTSRNRCQHCRLQKCLAVGMSRDAVKFGRMSKKSLA-
Mutant Genotyping Size
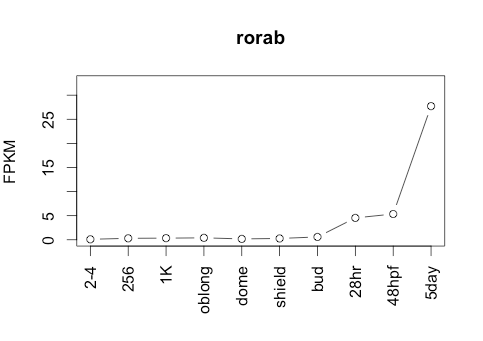
89Ribosome Profiling Development
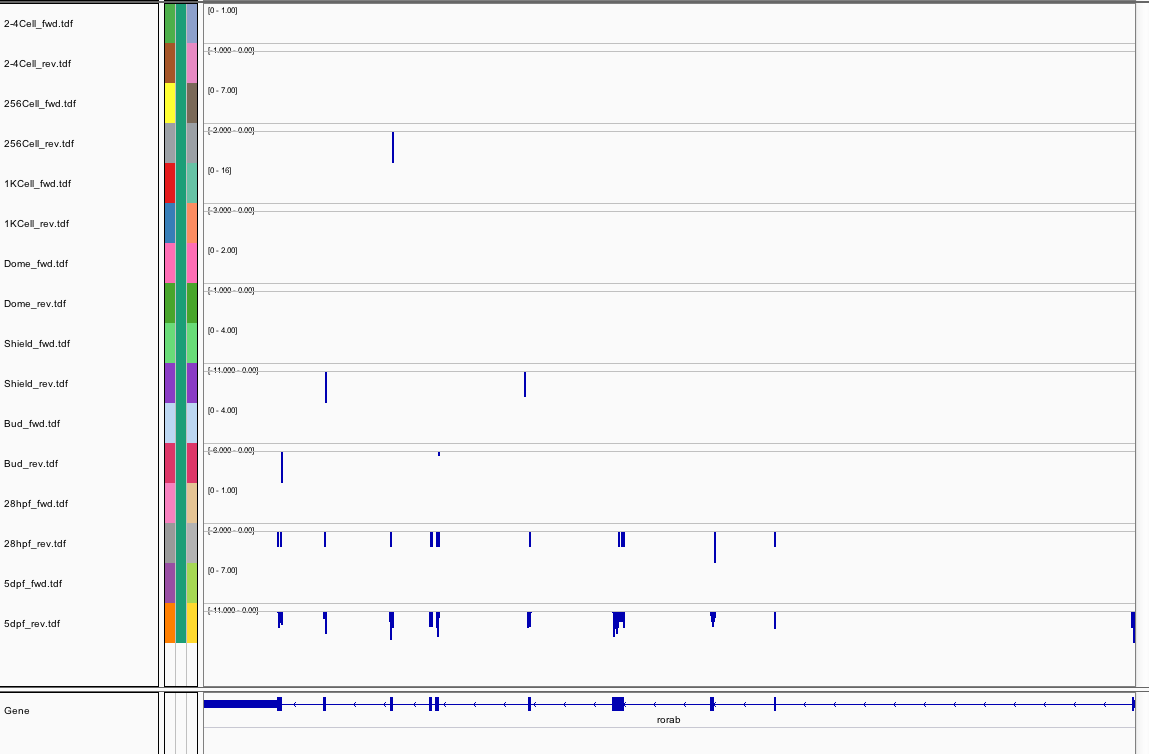
Transcript Plot
Behavior Data
Behavior Data Summary
NoneBehavior Data Description 1
Dark flash block 1 start Merged section Pvalue = 0.0268 27 hethom vs 18 homhom ribgraph_mean_ribbon_fullboutdata_dpix_a0darkflash103.pngBehavior Data Graph 1
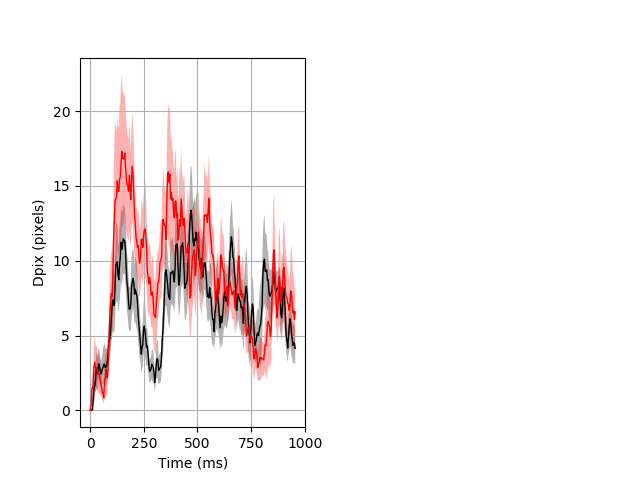
Behavior Data Description 2
Day all prepulse tap Merged section Pvalue = 0.0087 25 hethet vs 27 hethom ribgraph_mean_ribbon_fullboutdata_dpix_dayprepulseinhibition100d.pngBehavior Data Graph 2
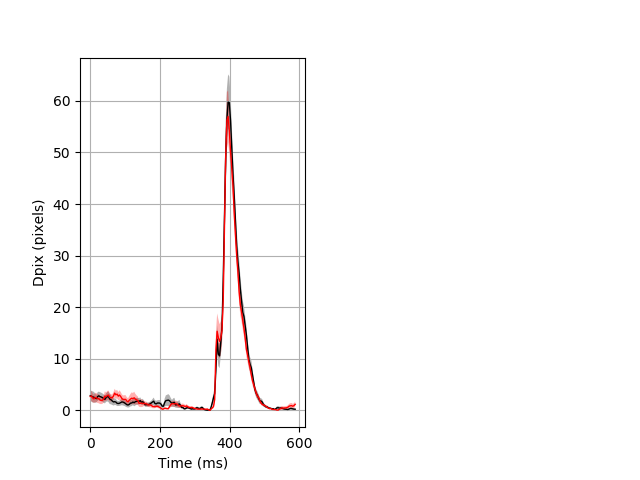
Behavior Data Description 3
Features of movement entire protocol Graph Pvalue = 0.000886 Merged section Pvalue = not significant 21 homhet vs 18 homhom ribgraph_mean_ribbonbout_aveboutdisp_10min_combo.pngBehavior Data Graph 3
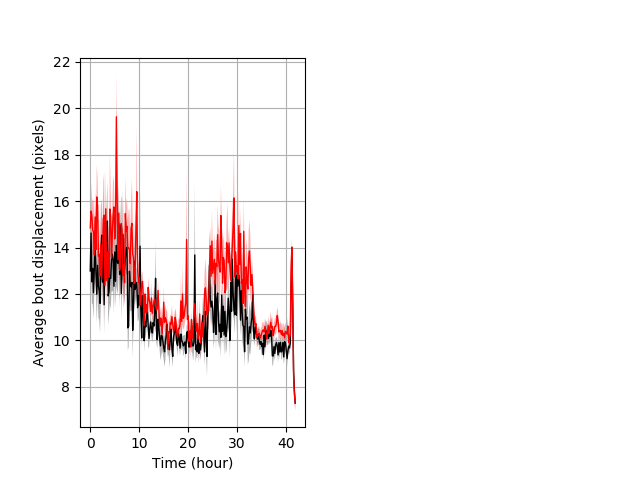
Behavior Data Description 4
Frequency of movement entire protocol Graph Pvalue = not significant Merged section Pvalue = not significant 25 hethet vs 18 homhom ribgraph_mean_ribbonbout_dpixnumberofbouts_10min_combo.pngBehavior Data Graph 4
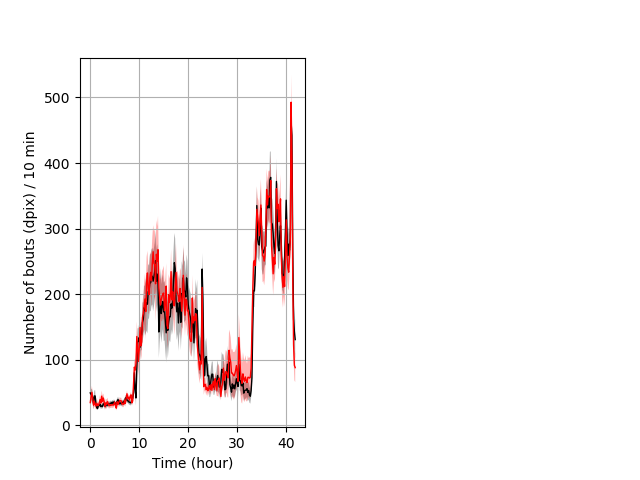
Behavior Data Description 5
Location in well entire protocol Graph Pvalue = not significant Merged section Pvalue = not significant 25 hethet vs 18 homhom ribgraph_mean_ribbonbout_averhofrac_10min_combo.pngBehavior Data Graph 5
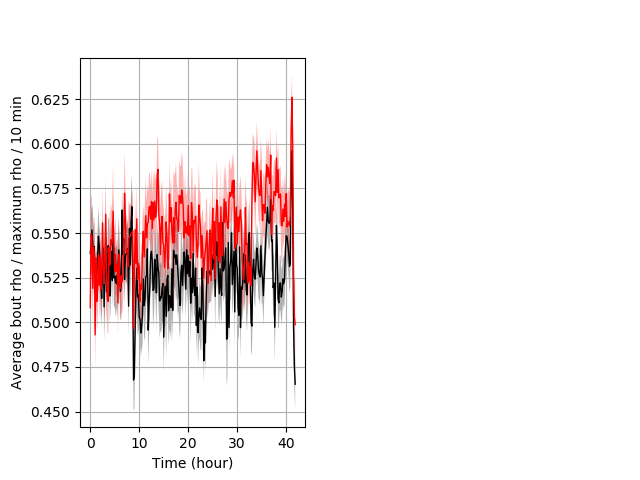
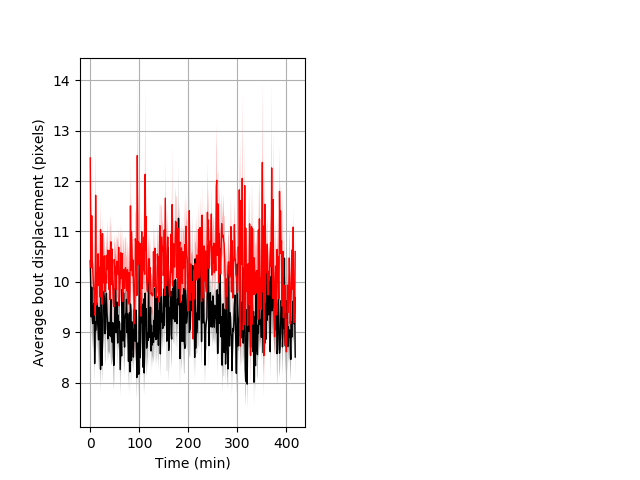
Behavior Data Description 6
Features of movement Graph Pvalue = 0.002 25 hethet vs 18 homhom ribgraph_mean_ribbonbout_aveboutdisp_min_day2darkflashes.pngBehavior Data Graph 6
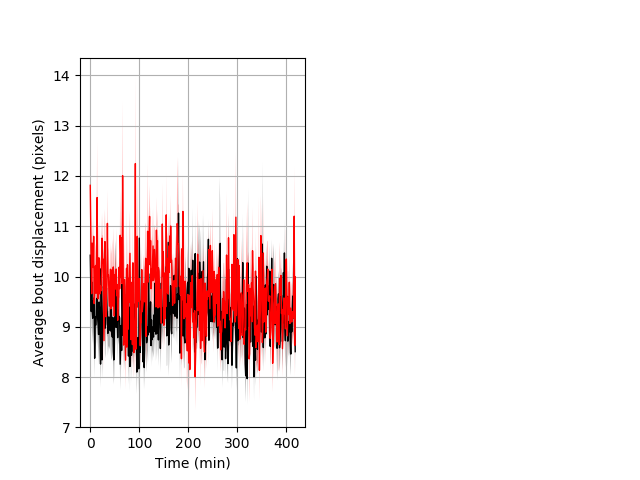
Behavior Data Description 7
Features of movement Graph Pvalue = 0.008 25 hethet vs 21 homhet ribgraph_mean_ribbonbout_aveboutdisp_min_day2darkflashes.pngBehavior Data Graph 7
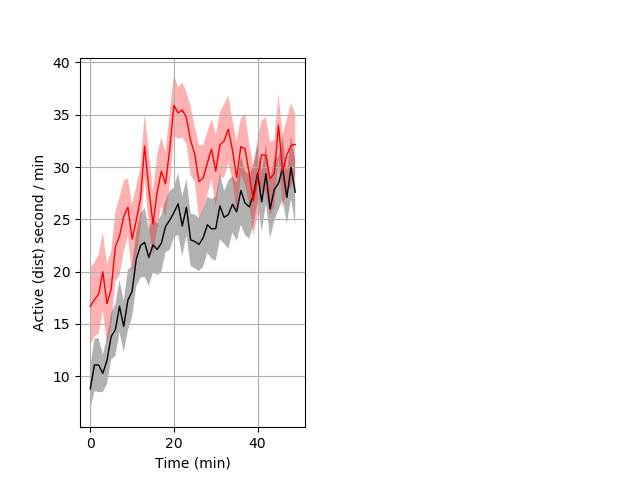
Behavior Data Description 8
Frequency of movement Graph Pvalue = 0.003 25 hethet vs 18 homhom ribgraph_mean_ribbon_distsecper_min_day2morning.pngBehavior Data Graph 8
Behavior Data Description 9
Location in well Graph Pvalue = 0.008 25 hethet vs 21 homhet ribgraph_mean_ribbonbout_interboutaverhofrac_10min_day2night1.pngBehavior Data Graph 9
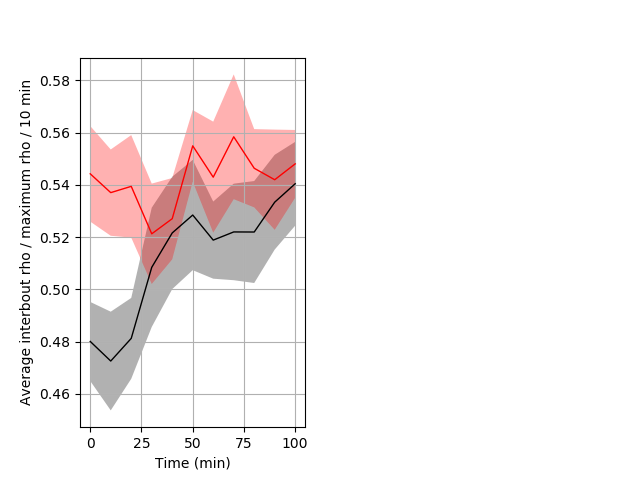
Behavior Data Description 10
Location in well Graph Pvalue = 0.02 25 hethet vs 18 homhom ribgraph_mean_ribbonbout_interboutaverhofrac_10min_day2night1.pngBehavior Data Graph 10
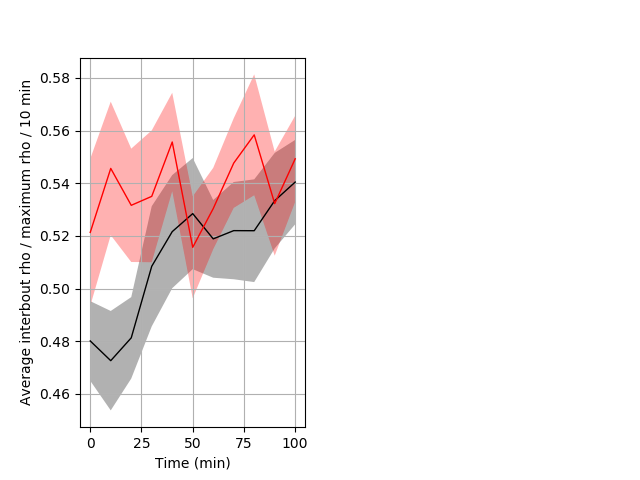
Behavior Data Description 11
NoneBehavior Data Graph 11
NoneBehavior Data Description 12
NoneBehavior Data Graph 12
NoneBehavior Data Description 13
NoneBehavior Data Graph 13
NoneBehavior Data Description 14
NoneBehavior Data Graph 14
None