Locus Rank
45Gene
Id
189Gene Name
VPS13CDuplicated
FalseMaternal
TrueGene Not Within Locus (Nearby
FalseGene Description
Mouse Phenotype
Additional Image
Allen Brain

Allen Regions
Zfin In Situ
Zfin Brain Areas
Published Zebrafish Pehnotype
Not In Allen Brain
TrueZFIN Link
http://zfin.org/ZDB-GENE-070426-4Allen Link
http://mouse.brain-map.org/gene/show/107463More Additional Images
Papers
http://www.researchgate.net/profile/Anthony_Monaco/publication/8219544_Analysis_of_the_human_VPS13_gene_family/links/0912f50dd91a40cfd7000000.pdfLocus Rank
Locus Rank
45Genes in Region
RORA, VPC13CAssociated Snps
rs12903146GWAS Region (hg19)
chr15:61831663-61909663Genes Skipping
VPS13C was made, but then the zebrafish genome changed and a new/dominant isoform was predicted that was not eliminated in the tested allele. Maybe worth remaking, although RORA has additional associations with schizophrenia in other studies, as well as associations with autism and depression.Ricopili Plot Surrounding Area
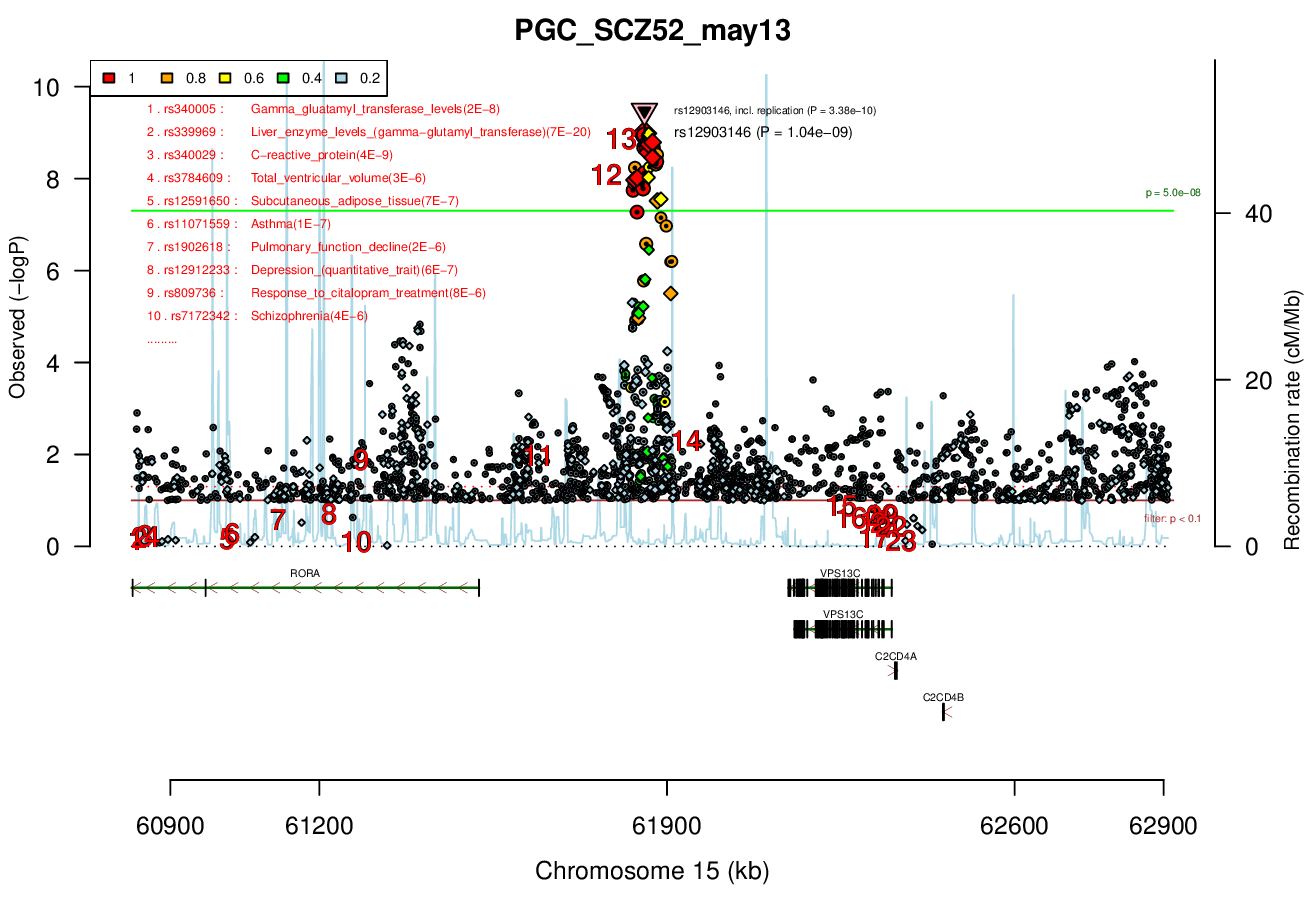
Distance On Each Side Of Locus In Above Plot
1 MBRicopili Plot
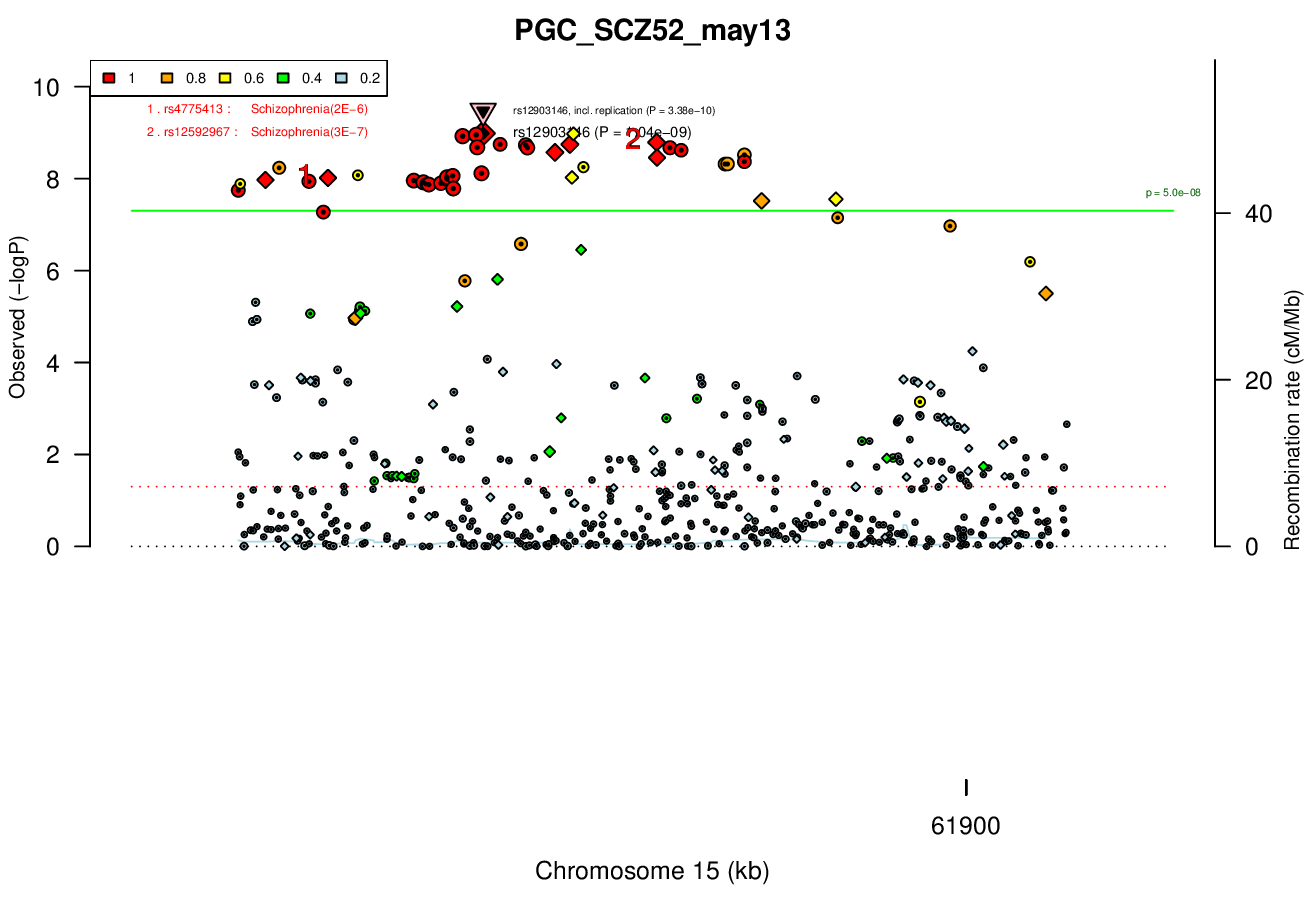
Gwas And Surrounding Region Pic
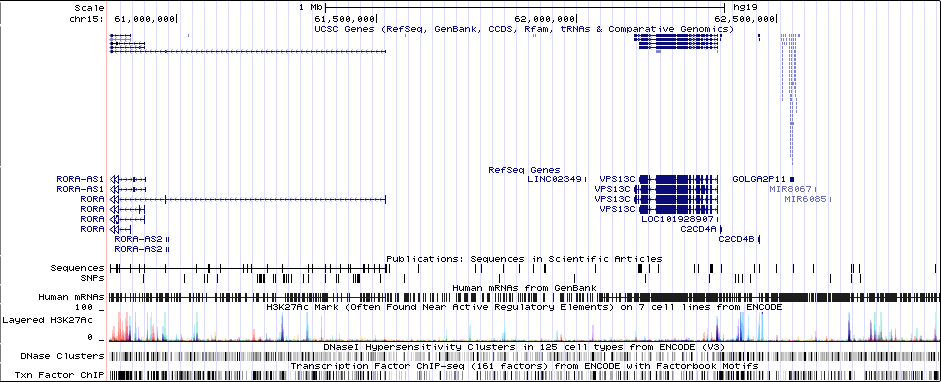
Gwas Region Pic
Sequence 1
Ensembl Gene Name
vps13cMutation Area - WT DNA Sequence
ggacggcagACTCTCAAGCTGGAGAGTTTGTGTTTAATCTGGAGAGTTATGTGTATAAGAATCCACCGCGGGgtatgtacagacccattgctatagaaggccacttgcgcccatcatacatttgtttgtttttgatagtttcattttaagtgtgtgttgagagtgttgatttgttttcctcagcactgcttggtgtgcttttgtgtacatttctgcgagtgtgtttttattgcaagtgtgcgtgtttaatacttttctattctttctcaactgacttttctcttttccccaacgctgcgctctgtaagACAAAGGACGTAAGGTTAAAAAGCATAAAAAACCCTTTAGGAGATCAAAAAAGTATGAACATAAAGCAGgtgggtggtcagaagcgactgtgatcgactcgtcacggcaaatattcacaatagttacattttatttgttagaataaaagcatttatttccatggtgctagattaagtcattagaatggcaaatgtaatggacttgcttataaaccctttgtatttaaaattttagAGAAGCCTCAGGAAGAAAAGAAGGACACGTTTGCTGAGAAACTGGCAACACAAGTTATCAAAAACCTCCAGGTCAAAATCACAAGCATTCACATCAGATATGAAGATGATgtaagtgttgacttgtcaaacatgatgtcagctcttcagttatgtgcatttcttgaatgtttgcatgaaataatgataaggatgcagcttttatttgatggtatatttattaatggtatattgtgcctcagGTGTCCGATCCTCAAAAACCTCTTGCTATGGGTTTGACCCTTTCAGAACTCAGCCTACAGgMutation Area - Mutant DNA Sequence
ggacggcagACTCTCAAGCTGGAGAGTTTGTGTTTAATCTGGAGAGTTATGTGTATAAGAATCGCGGGgtatgtacagacccattgctatagaaggccacttgcgcccatcatacatttgtttg
This mutant is likely not a true null because it only removes one of two predicted isoforms. When the gRNAs were designed, the zebrafish genome only predicted one isoform.
Guide RNA Target Sites
GTGTATAAGAATCCACCGCGGGg
TTTTGAGGATCGGACACctgagg
ATGAACATAAAGCAGgtgggtgg
Tcttacagagcgcagcgttgggg
Genotyping Primers
f, ggacggcagACTCTCAAGCTGGAG
r, caaacaaatgtatgatgggcgcaagtgg
Alignment of WT and Mutant Sequences w/ gRNA Targets (cyan) and Genotyping Primers (pink) Shown on WT Sequence

Allele
4bpDWT Genotyping Size
128WT Protein Sequence
MVFESLVSDLLNRFIGDYVENLDKSQLKIGIWGGNVVLENLKVKENALSELDVPFKVKAG
QIGKLTLKIPWKNLYNEAVVATLDGLYLLVVPGAIESKLFSSSYNAIKYDAVKEERYLQE
AKQKELQRIEETLQLVARRDSQAGEFVFNLESYVYKNPPRDKGRKVKKHKKPFRRSKKYE
HKAEKPQEEKKDTFAEKLATQVIKNLQVKITSIHIRYEDDVSDPQKPLAMGLTLSELSLQ
TADENWKTCILNEAAKIIYKLGRLECFCAYWNVNGPIFYKDSWEGIVEKLKAGISTKNEE
LQGYQYIFKPIFASARMCINPNAEVELKSPKANLHLEVQNIAIEMTKSQYLSMVDLLESI
DCMVKNGPYRKFRPDVPVHRNARQWWKYGISSILEVHVRRFNQMWNWTNIKKHRQTLKSY
KAAYKVKLTQSAKVREDTEKQIQELEKGLDVFNIVLARQQVQMEVVRSGQKLVAKKAATQ
KQSGGGFFSSFFGKKEGKKEEEDKEPESIDSIMTVEEKSKLYTAIGYSGSSHNLALPKHY
VAVIVNFKLLSTSVTFREEPGVPEILKVQMIDLSTSISQRPGAQAIRVEAKLEHWYVTGL
QQQGEVPSLIASLGDSNSSLLSVRFEINPEDSPADQLLRVHSQPVEIIYDALTVISITEF
FKTGKGVDLEVITSATLSKLEEIKEKTATGLSHIIETRKVLDLRIDLKPSYLLLPKSGFY
DGKSELMIVDFGYLQLNSVDQGGHQQVSASFSSLEEIMDRAYERYSLELRSVQILYSRSG
EKWKSACHEGLSHQHILQPMDFTLQLAKCMVDKDARMPRFKVSGELPLLHLKISDQKIQG
VLELVDSIPLPQTSSSPPSSPTGKKVLGMPLSNTRPKVLALDPTVLSYAVESESDEDTSE
KSVDEESLRSTLEELTEVQFQFEVKAVLFELTRQAAQEDVVLAFSVSQLGAEGRIRTFDL
SVTSYLKKISLDYCEEKDVQNQPLHLITSSDKHGSDLLKVEYIKADVNGPSFCTLFNNTE
QTLKVEFSSLDFLLHTKALLSTINYMNSAIPQELTANKERETRRRADRAAAGKTVSKGSK
DAGVVNFKLSAVLGSFRVAVCDDRSNIADIRVQGIDASVMVQAKETDVFARLRDIVVLDV
DPKTIHKKAVSIVGEEVFSFKMTLYPGATEGDGYTDTSKVDGKVIMRLGCIQIVYLHKFL
MSLLMFVDNFQTAKEALSAATAQAAEKAASSVRDFAQKSFRLAMDIRLKAPLIIIPQSST
SHNAFVVDLGLITVGNRFSLLAAEGFPLPAVLETMDIKLTQLKLSRTVLKHDADHLDIEI
LEPINLELLVKRNLAAAWYNKIPGVEVNGVLMSMNMSLGQEDLGVLMRILAENIGEGSKA
AAVEGTKASAKDEAVIDEGLMQTLPVGIGSLPALTNGAFTENVVNVLLNFEIKKVMLMLK
KSKNGDESPFLVLDVSQLGIDTKVRKYDMEATTYIKTISMKCLEFPDSNGEPLCIISSSA
ESGADLLEVKYFKADRNGPNFASLYQNTEQKMNVVFSSLDLMLHTEALLSTMDFLSAALS
TSSLPSPERESKKSEDMKTTSAKSTALSSPSDGDIIDLMVNMQLGAFNVLVCDQKSNMAD
IKIQGLDGSLQMKGTQTHMFTRLRDFIVINVDSKTIHKKAISIVGDEVFSFSMSLTPKAT
EGAGYTDTSKVDGKVKLNVGCIQVVYLHKFFMSLLNFSNNFQEAKEALSAATAQAAEKAA
SSVRDFAQKSFRLAMDIKLKAPLIIIPQSSTSHNAVEADLGLITVGNRFSLLPVEGRPLP
AVIDNMDIQLTQLKLSRISMELDSDQTSSKLLEPVNLVLSVKRNLASSWYQKMAAIEVDG
DLKPMKVALSQDDLSVLLRILMENIGEASDIQSDTALRQENVLVRARTATGSEILEEKLS
ESKLKETELLESVRFSFNVESLGLTLYNNDPSQPCVHEEKSCLGEFMLCKMKMSGKMLNN
GNMEVSTILTTCTLDDRRSGIQRVTSRMLGRRDEETSEPMIDVTYSQSVDERSVVAVLQK
LYLCASVEFLLAVADFFVQALPTSPPTQREKTNQLPLKYVSEPKIHTEPKAVPAQRTKLR
AVVVDPEVVFVANLMKADAPALVASFQCDFSLLSEEAGQTMKANVRSLKVLACPFIRAEG
DKAVTTVLRPCSVELETKMPANAPLAGSMTVEEVIVKISPIILNTVITITAAMAPKPKEE
LSQQKSGDQNSLWSLMSVANANYWFLGVDTASEVTESFTDVDSSKDGETFGLTVKVVQVT
LESGLGHRTVPLLLAESSFSVTAQNWSSLLSVSADMTLEVNYFNETHAVWEPLLERVDNG
KRRWNLKLDMKNNPVRDKSPVPGDDFIMLPDPRTAITICSKDTMNITVSKCCLNVFNNLA
KAFSESTASTFDHSLKEKSPFTIRNALGVALIVQHSANLRVPGSSTLGKVHELAVDKSLD
LEYSSFERSSRQSALQRQESCLFNLSIVPSGYSEISNIPLGKPGRRLYNVRQPMQEESVS
VLLQIDASEGNKVITVRSPLQIKNHFSVPFAILKYSSELGGMVNVGVAEPEKECHVPLEA
YRSQLFLLPVGPLEKLYRASTTCIAWKEKVHVSSEVHAVLQCPATESNFLPLVVNTLAMP
DDLSHIANHGEVEWDPAYLIHLHPPVTVKNLLPYSIRYLLEGSADIYELQEGSAADVLNS
NVSGEIMELILIKYQGRNWTGRMRISRDMPEFITICFNCDTGEALTVDVCVHVSRVSGKF
ILSVHSPYWIINKTSRVLQYRAEDTFVKHPSDFRDVILFSFKKKNIFSKNKLQLCVSTSS
WSEGFSLDTVGSYGCVRCPDKSGDYLVGVSIQMSSFNLTRIVTMSPFYTLVNKSLFELEV
GEVQNNNGSSNKWHYISSTECLPFWPESSTEKLCVRVVGCESSSKSFFFNKQDSGTLLRM
DQYGGVIVDVNISDHSTVISFTDYYDGAAPALVVNHTPSATVNFRQRGCEECWALRPGEA
QRFAWDDPAGVRKLCWSCQDHSGELDLVKDECGQFAYSSLVQIHWVSFLDGRQRVLIFTE
DVGIVTKARQAEELEQFQQEVNVSLQNLGLSLINNDIRQEIAYVGITSSGVVWEMRPKNR
WKSFNHKNIGLLEKAYQNHINKSDPGWTMLETGIEVNFGRVPMLMRQPFSCQIRRNFLPG
IHVELKQSPHQRSLRAQLYWLQVDNQLPGAIFPTVFHPVPLPKSIVQDSEPKPFIDVSII
TRFNEHSQVMQFKYFMALVQEMAVKIDQGFLGAILALFTPATDSLDSKKKNKLIQKDLDA
LQAQLMESSLTDTSGLSFFEHFHISPIKLHLSLSLGSSGDESEQGDMVAIQSVNLLLKSI
GATLTDVDDLIFKLAYFEVKYQFYRRDKLMWAVIRHYTEQFLTQMYVLVLGLDVLGNPFG
LIRGLSEGVEAFFYEPIQGAVQGPEEFAEGLVIGVRSLLGHTVGGAAGMVSRITGSVGKG
LAAITMDKEYQQKRREEMNRPTKDFGESLAKGGKGFLKGVVGGVTGIVTKPVEGAKKEGA
AGFFKGIGKGLVGVVARPTGGIVDMASSTFQGIQRVAESTEEVTKLRPVRLIHEDGIIRP
YDHHASEGYDLFQPEEM-
Mutant Protein Sequence
MVFESLVSDLLNRFIGDYVENLDKSQLKIGIWGGNVVLENLKVKENALSELDVPFKVKAG
QIGKLTLKIPWKNLYNEAVVATLDGLYLLVVPGAIESKLFSSSYNAIKYDAVKEERYLQE
AKQKELQRIEETLQLVARRDSQAGEFVFNLESYVYKNRGTKDVRLKSIKNPLGDQKSMN
IKQRSLRKKRRTRLLRNWQHKLSKTSRSKSQAFTSDMKMMCPILKNLLLWV-
Mutant Genotyping Size
124Behavior Data
Behavior Data Summary
NoneBehavior Data Description 1
NoneBehavior Data Graph 1
NoneBehavior Data Description 2
NoneBehavior Data Graph 2
NoneBehavior Data Description 3
NoneBehavior Data Graph 3
NoneBehavior Data Description 4
NoneBehavior Data Graph 4
NoneBehavior Data Description 5
NoneBehavior Data Graph 5
NoneBehavior Data Description 6
NoneBehavior Data Graph 6
NoneBehavior Data Description 7
NoneBehavior Data Graph 7
NoneBehavior Data Description 8
NoneBehavior Data Graph 8
NoneBehavior Data Description 9
NoneBehavior Data Graph 9
NoneBehavior Data Description 10
NoneBehavior Data Graph 10
NoneBehavior Data Description 11
NoneBehavior Data Graph 11
NoneBehavior Data Description 12
NoneBehavior Data Graph 12
NoneBehavior Data Description 13
NoneBehavior Data Graph 13
NoneBehavior Data Description 14
NoneBehavior Data Graph 14
None