Locus Rank
27Gene
Id
170Gene Name
SLC32A1Duplicated
FalseMaternal
FalseGene Not Within Locus (Nearby
FalseGene Description
Mouse Phenotype
Homozygous null mice been independently reported to die perinatally exhibiting a hunched posture, respiratory failure, cleft secondary palate due to failure of palate shelf elevation, umbilical hernia or omphalocele, and loss of neurotransmitter release in both GABAergic and glycinergic neurons.Additional Image
Allen Brain
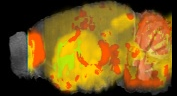
Allen Regions
Zfin In Situ
Zfin Brain Areas
Published Zebrafish Pehnotype
Not In Allen Brain
FalseZFIN Link
http://zfin.org/ZDB-GENE-061201-1Allen Link
http://mouse.brain-map.org/experiment/show/72081554More Additional Images
Papers
http://www.ncbi.nlm.nih.gov/pubmed/12638124Locus Rank
Locus Rank
27Genes in Region
ACTR5 PPP1R16B SLC32A1 FAM83D DHX35Associated Snps
rs6065094GWAS Region (hg19)
chr20:37361494-37485994Genes Skipping
ACTR5 - not in brain at all, DNA repair; FAM83D - mitosis, cancer, not in zebrafish brain; DHX35 - likely RNA helicase, not in allen brainRicopili Plot Surrounding Area
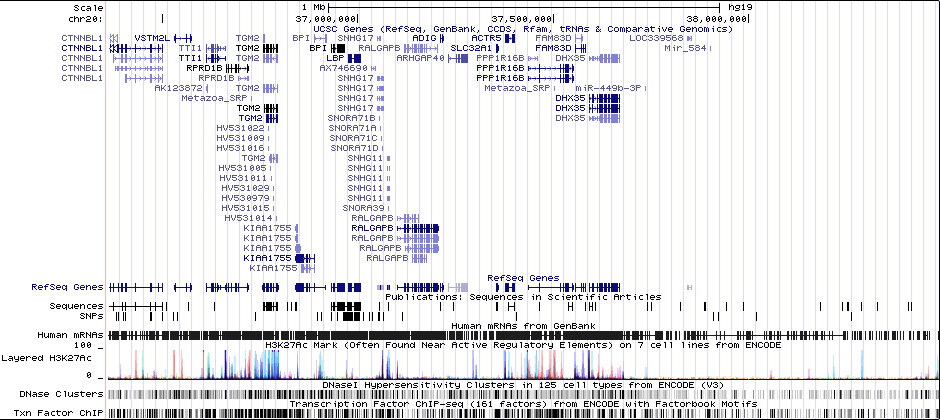
Distance On Each Side Of Locus In Above Plot
1 MBRicopili Plot
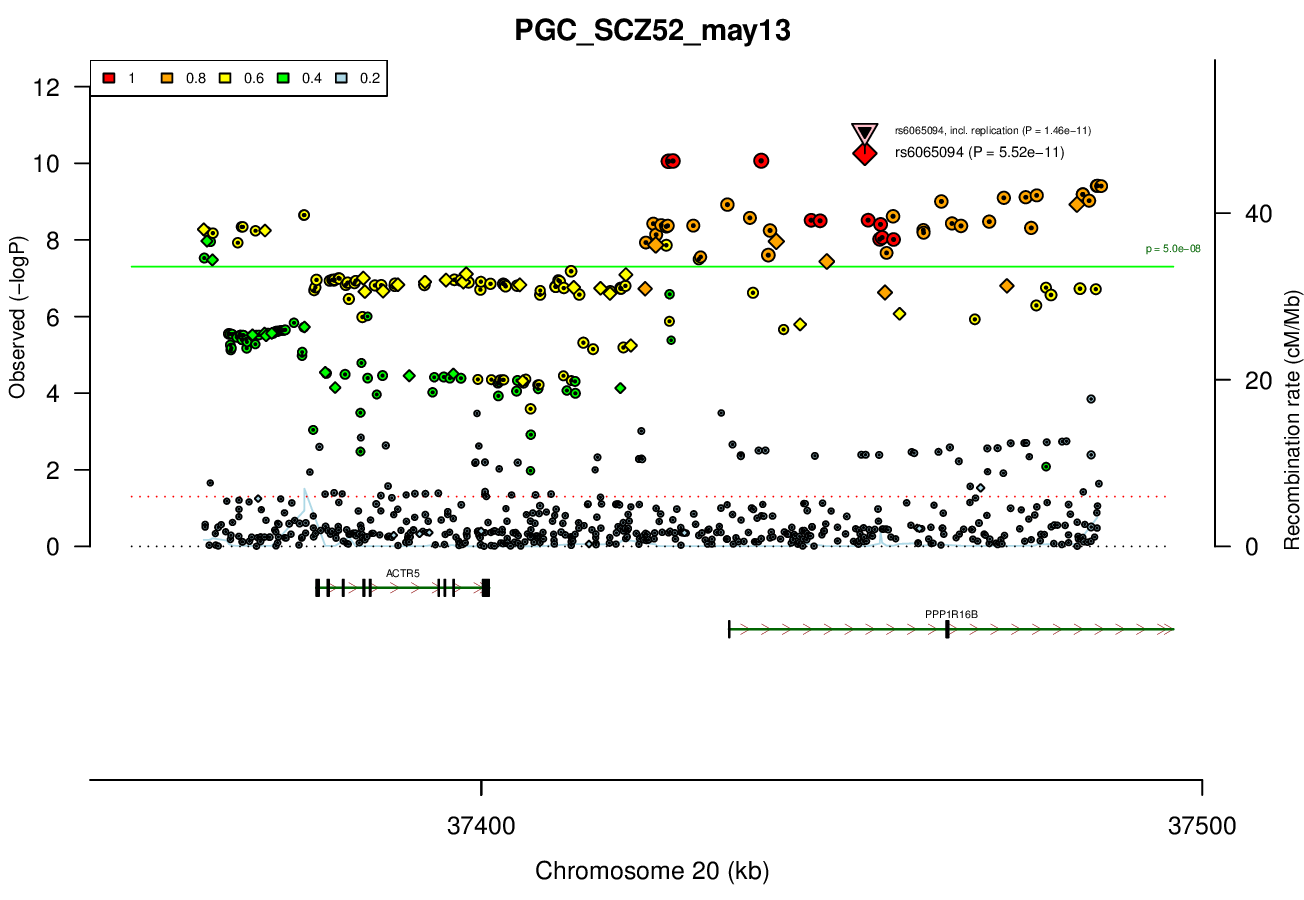
Gwas And Surrounding Region Pic
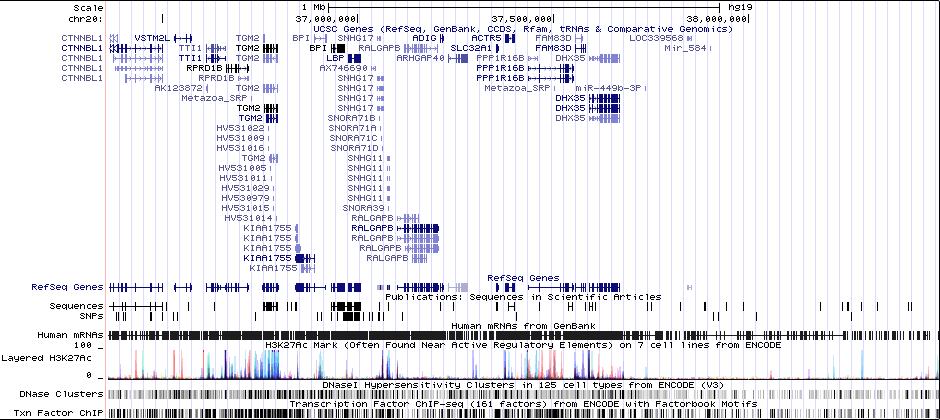
Gwas Region Pic
Sequence 1
Ensembl Gene Name
slc32a1Harvard Allele
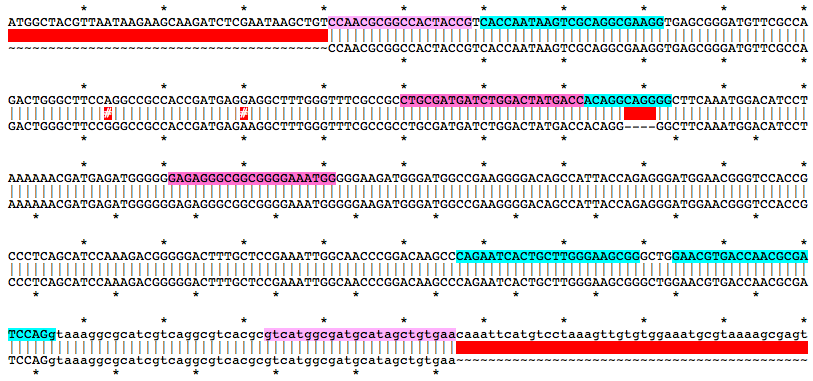
a332Mutation Area - WT DNA Sequence
CCAACGCGGCCACTACCGTCACCAATAAGTCGCAGGCGAAGGTGAGCGGGATGTTCGCCAGACTGGGCTTCCAGGCCGCCACCGATGAGGAGGCTTTGGGTTTCGCCGCCTGCGATGATCTGGACTATGACCACAGGCAGGGGCTTCAAATGGACATCCTAAAAAACGATGAGATGGGGGGAGAGGGCGGCGGGGAAATGGGGGAAGATGGGATGGCCGAAGGGGACAGCCATTACCAGAGGGATGGAACGGGTCCACCGCCCTCAGCATCCAAAGACGGGGGACTTTGCTCCGAAATTGGCAACCCGGACAAGCCCAGAATCACTGCTTGGGAAGCGGGCTGGAACGTGACCAACGCGATCCAGgtaaaggcgcatcgtcaggcgtcacgcgtcatggcgatgcatagctgtgaaMutation Area - Mutant DNA Sequence
CCAACGCGGCCACTACCGTCACCAATAAGTCGCAGGCGAAGGTGAGCGGGATGTTCGCCAGACTGGGCTTCCGGGCCGCCACCGATGAGAAGGCTTTGGGTTTCGCCGCCTGCGATGATCTGGACTATGACCACAGGGGCTTCAAATGGACATCCTAAAAAACGATGAGATGGGGGGAGAGGGCGGCGGGGAAATGGGGGAAGATGGGATGGCCGAAGGGGACAGCCATTACCAGAGGGATGGAACGGGTCCACCGCCCTCAGCATCCAAAGACGGGGGACTTTGCTCCGAAATTGGCAACCCGGACAAGCCCAGAATCACTGCTTGGGAAGCGGGCTGGAACGTGACCAACGCGATCCAGgtaaaggcgcatcgtcaggcgtcacgcgtcatggcgatgcatagctgtgaaGuide RNA Target Sites
CACCAATAAGTCGCAGGCGAAGG
TGGACTATGACCACAGGCAGGGG
CAGAATCACTGCTTGGGAAGCGG
GAACGTGACCAACGCGATCCAGg
Genotyping Primers
f, CTGCGATGATCTGGACTATGACC
r, CCATTTCCCCGCCGCCCTCTC
Alignment of WT and Mutant Sequences w/ gRNA Targets (cyan) and Genotyping Primers (pink) Shown on WT Sequence
Allele
4bpDWT Genotyping Size
92WT Protein Sequence
MATLIRSKISNKLSNAATTVTNKSQAKVSGMFARLGFQAATDEEALGFAACDDLDYDHRQ
GLQMDILKNDEMGGEGGGEMGEDGMAEGDSHYQRDGTGPPPSASKDGGLCSEIGNPDKPR
ITAWEAGWNVTNAIQGMFVLGLPYAILHGGYLGLFLIIFAAVVCCYTGKILIACLYEENE
DGQLVRVRDSYVDIANACCAPRFPALGGHVVNVAQIIELVMTCILYVVVSGNLMYNSFPT
LPVSQRSWAIIATAALLPCAFLKNLKAVSKFSLLCTLAHFVINILVIAYCLSRARDWAWD
KVKFYIDVKKFPISIGIIVFSYTSQIFLPSLEGNMQKPSEFHCMMNWTHIAACILKGLFA
LVAYLTWADETKEVITDNLPSSIRAVVNLFLVSKALLSYPLPFFAAVEVLEKTFFQDGGR
AFFPDCYGGDGRLKSWGLSLRCALVVFTMLMAIYVPHFALLMGLTGSLTGAGLCFLLPSL
FHLKLLWRKLLWHQVFFDVAIFVIGGICSISGFIHSVEGLIEAYKYNLPD-
Mutant Protein Sequence
MATLIRSKISNKLSNAATTVTNKSQAKVSGMFARLGFRAATDEKALGFAACDDLDYDHRGFKWTS-Mutant Genotyping Size
88Ribosome Profiling Development
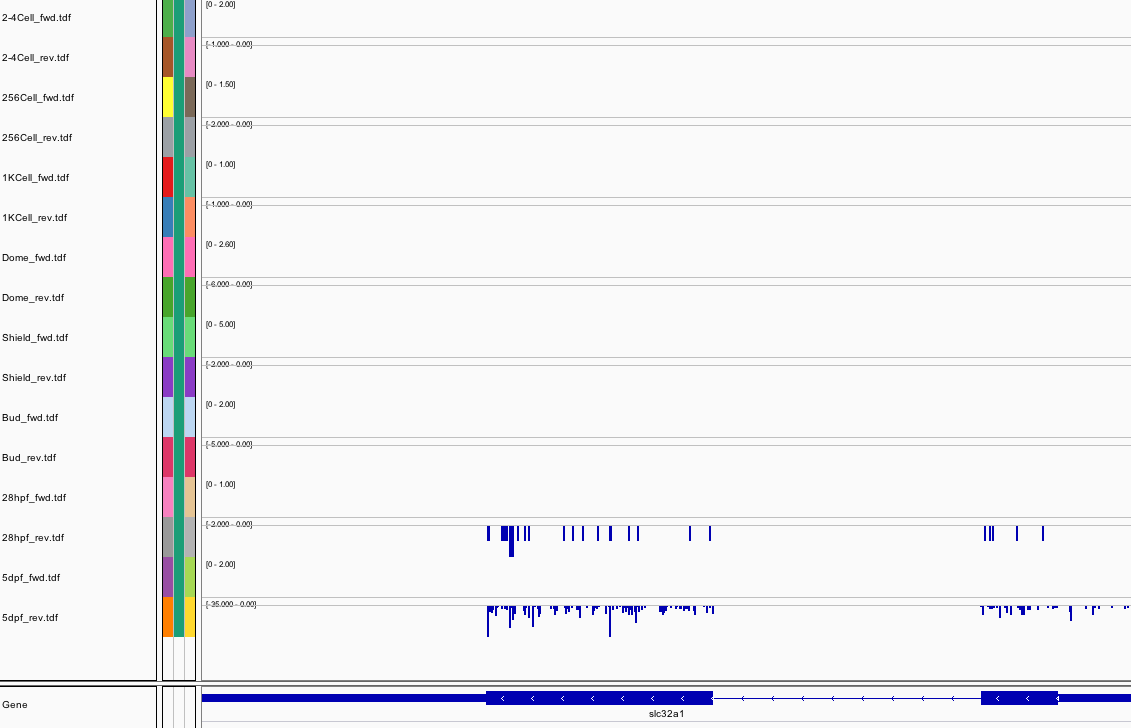
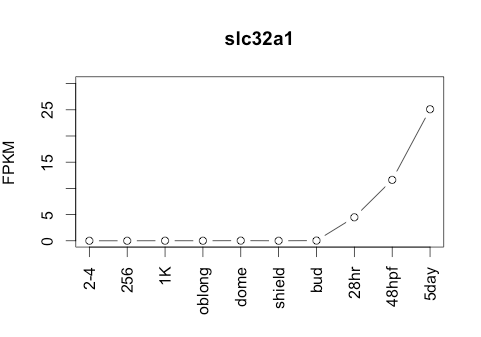
Transcript Plot
Behavior Data
Behavior Data Summary
NoneBehavior Data Description 1
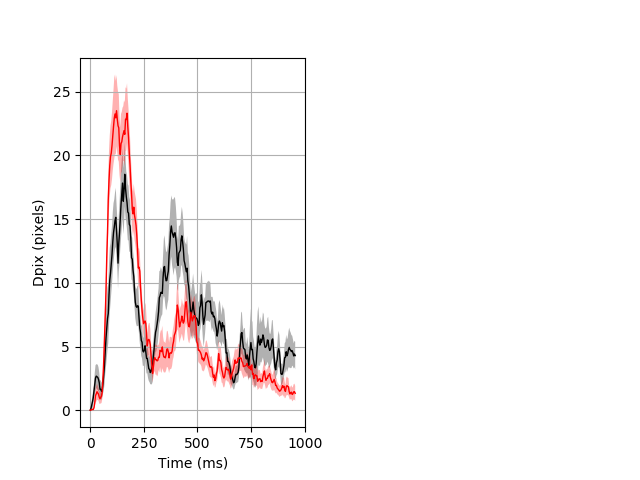
Dark flash block 1 start Merged section Pvalue = not significant 26 wt vs 35 het ribgraph_mean_ribbon_fullboutdata_dpix_a0darkflash103.pngBehavior Data Graph 1
Behavior Data Description 2
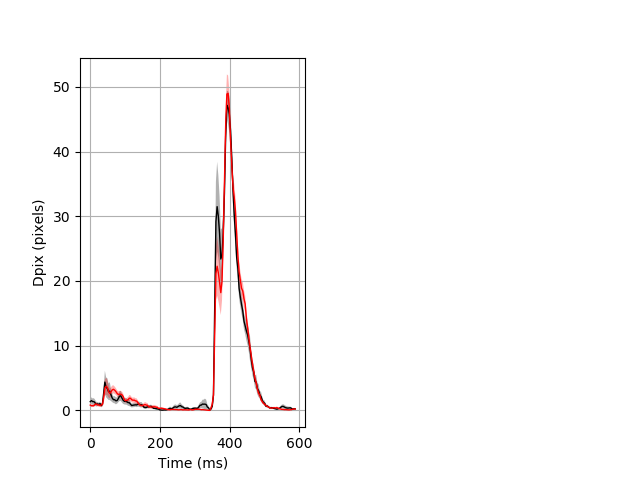
Day all prepulse tap Merged section Pvalue = not significant 26 wt vs 35 het ribgraph_mean_ribbon_fullboutdata_dpix_dayprepulseinhibition100d.pngBehavior Data Graph 2
Behavior Data Description 3
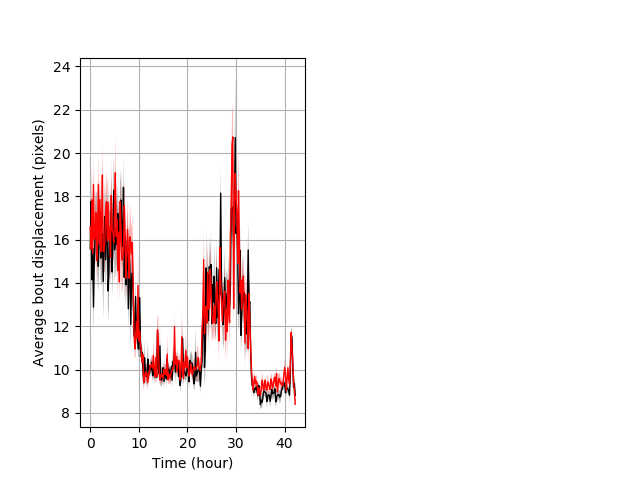
Features of movement entire protocol Graph Pvalue = not significant Merged section Pvalue = not significant 33 wt vs 40 het ribgraph_mean_ribbonbout_aveboutdisp_10min_combo.pngBehavior Data Graph 3
Behavior Data Description 4
Frequency of movement entire protocol Graph Pvalue = not significant Merged section Pvalue = not significant 26 wt vs 35 het ribgraph_mean_ribbonbout_dpixnumberofbouts_10min_combo.pngBehavior Data Graph 4
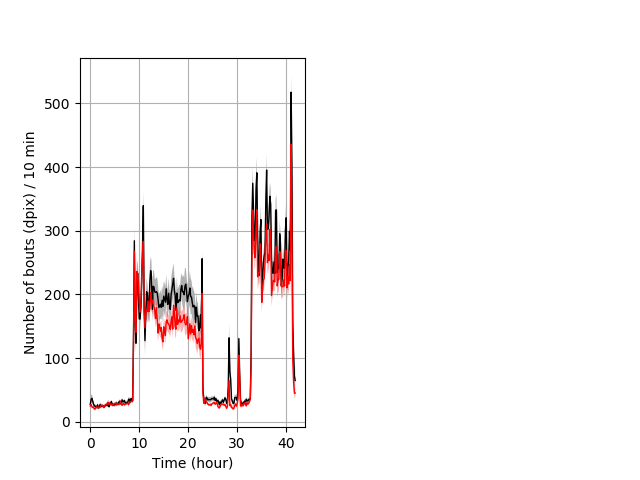
Behavior Data Description 5
Location in well entire protocol Graph Pvalue = not significant Merged section Pvalue = not significant 26 wt vs 35 het ribgraph_mean_ribbonbout_averhofrac_10min_combo.pngBehavior Data Graph 5
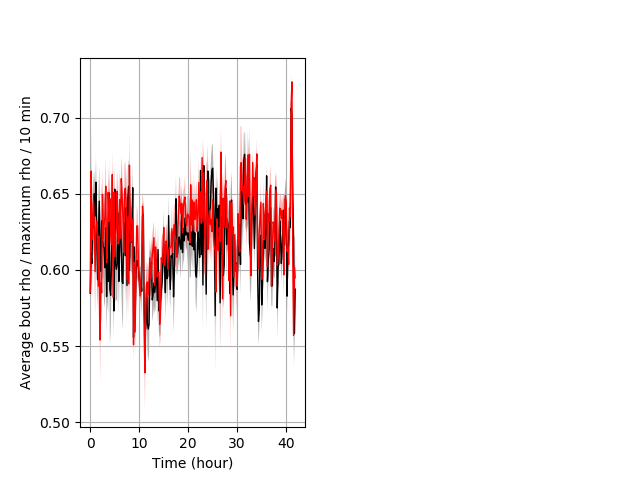
Behavior Data Description 6
Tap habituation Graph Pvalue = 0.0179 26 wt vs 35 het boxgraph_pd__ribgraph_mean_ribbon_slowfullboutdatamax_dist_cdaytaphab1.pngBehavior Data Graph 6
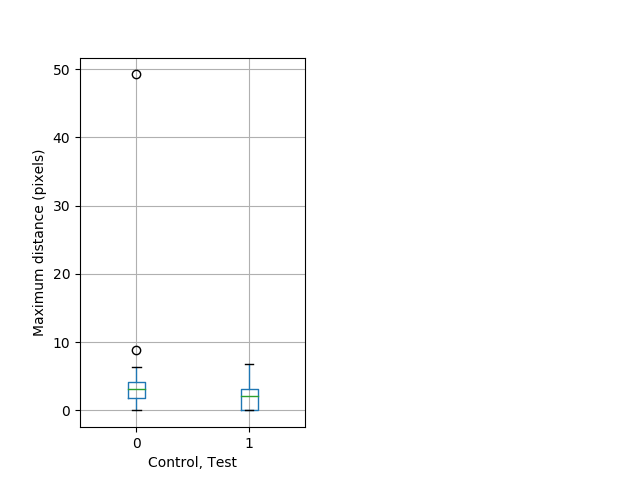
Behavior Data Description 7
NoneBehavior Data Graph 7
NoneBehavior Data Description 8
NoneBehavior Data Graph 8
NoneBehavior Data Description 9
NoneBehavior Data Graph 9
NoneBehavior Data Description 10
NoneBehavior Data Graph 10
NoneBehavior Data Description 11
NoneBehavior Data Graph 11
NoneBehavior Data Description 12
NoneBehavior Data Graph 12
NoneBehavior Data Description 13
NoneBehavior Data Graph 13
NoneBehavior Data Description 14
NoneBehavior Data Graph 14
None