Locus Rank
38Gene
Id
182Gene Name
SEZ6L2Duplicated
FalseMaternal
FalseGene Not Within Locus (Nearby
FalseGene Description
Mouse Phenotype
Additional Image
Allen Brain
Allen Regions
Zfin In Situ
Zfin Brain Areas
Published Zebrafish Pehnotype
Not In Allen Brain
FalseZFIN Link
http://zfin.org/ZDB-GENE-091204-189Allen Link
http://mouse.brain-map.org/gene/show/87745More Additional Images
Papers
Locus Rank
Locus Rank
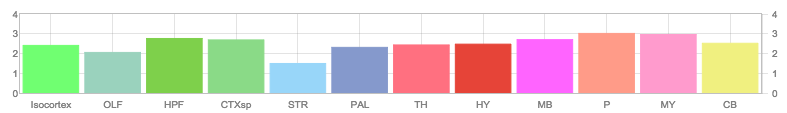
38Genes in Region
ALDOA ASPHD1 C16orf92 DOC2A FAM57B GDPD3 HIRIP3 INO80E KCTD13 MAPK3 PPP4C SEZ6L2 TAOK2 TBX6 TMEM219 YPEL3Associated Snps
rs12691307GWAS Region (hg19)
chr16:29924377-30144877Genes Skipping
ALDOA - definitely not; ASPHD1 - not in allen brain, and very unlikely; C16orf92 - nothing is known; FAM57B - hard to know, very little is known; GDPD3 - nothing in allen brain; HIRIP3 - histones; INO80E - maybe, but seems not that likely; MAPK3 - maybe; PPP4C - maybe, very unlikely; TAOK2 - maybe, but not in allen brain; TBX6 - not in allen brain, involved in development; TMEM219 - seems very unlikely, not in allen brain; YPEL3 - no.Ricopili Plot Surrounding Area
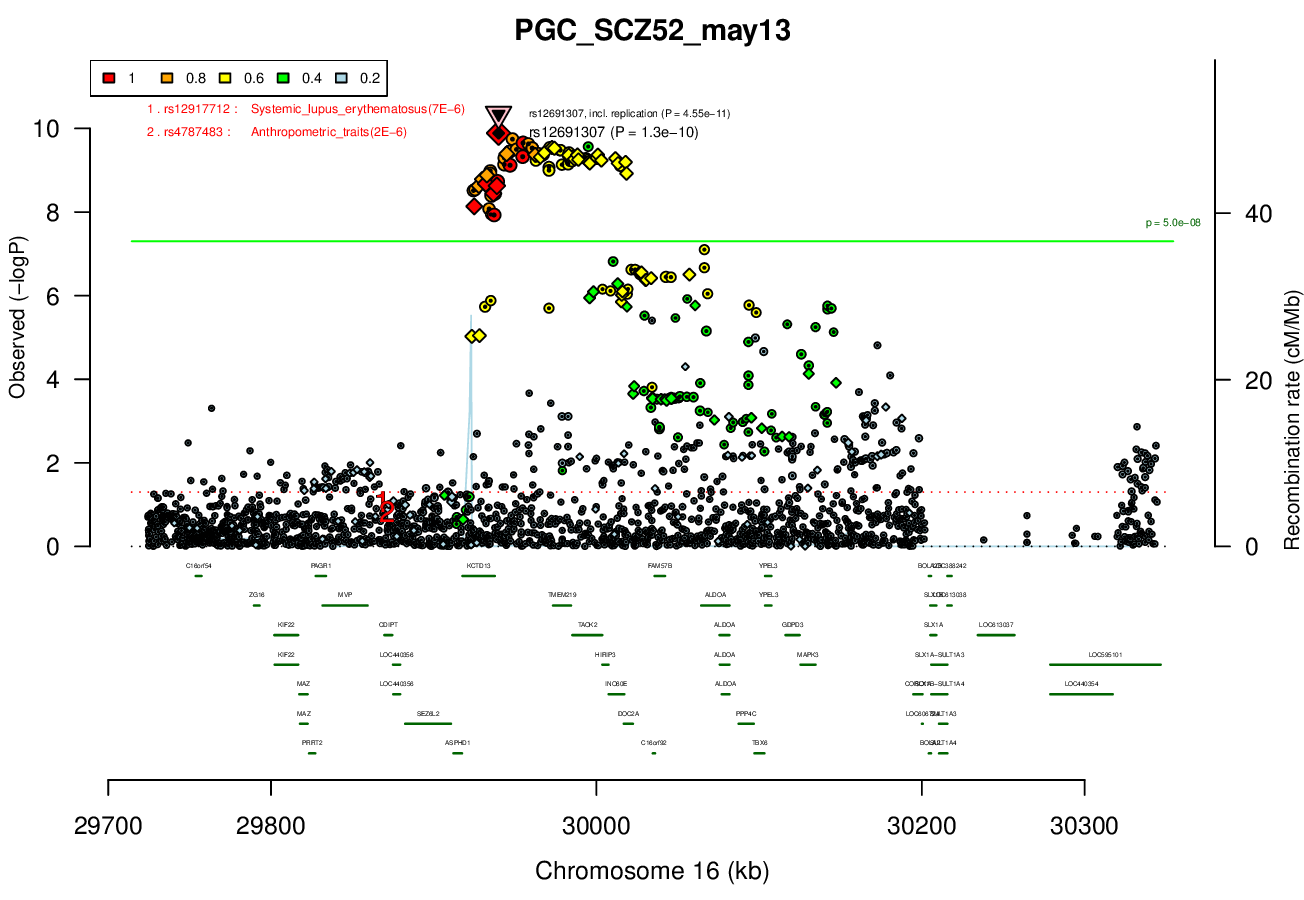
Distance On Each Side Of Locus In Above Plot
0.2 MBRicopili Plot
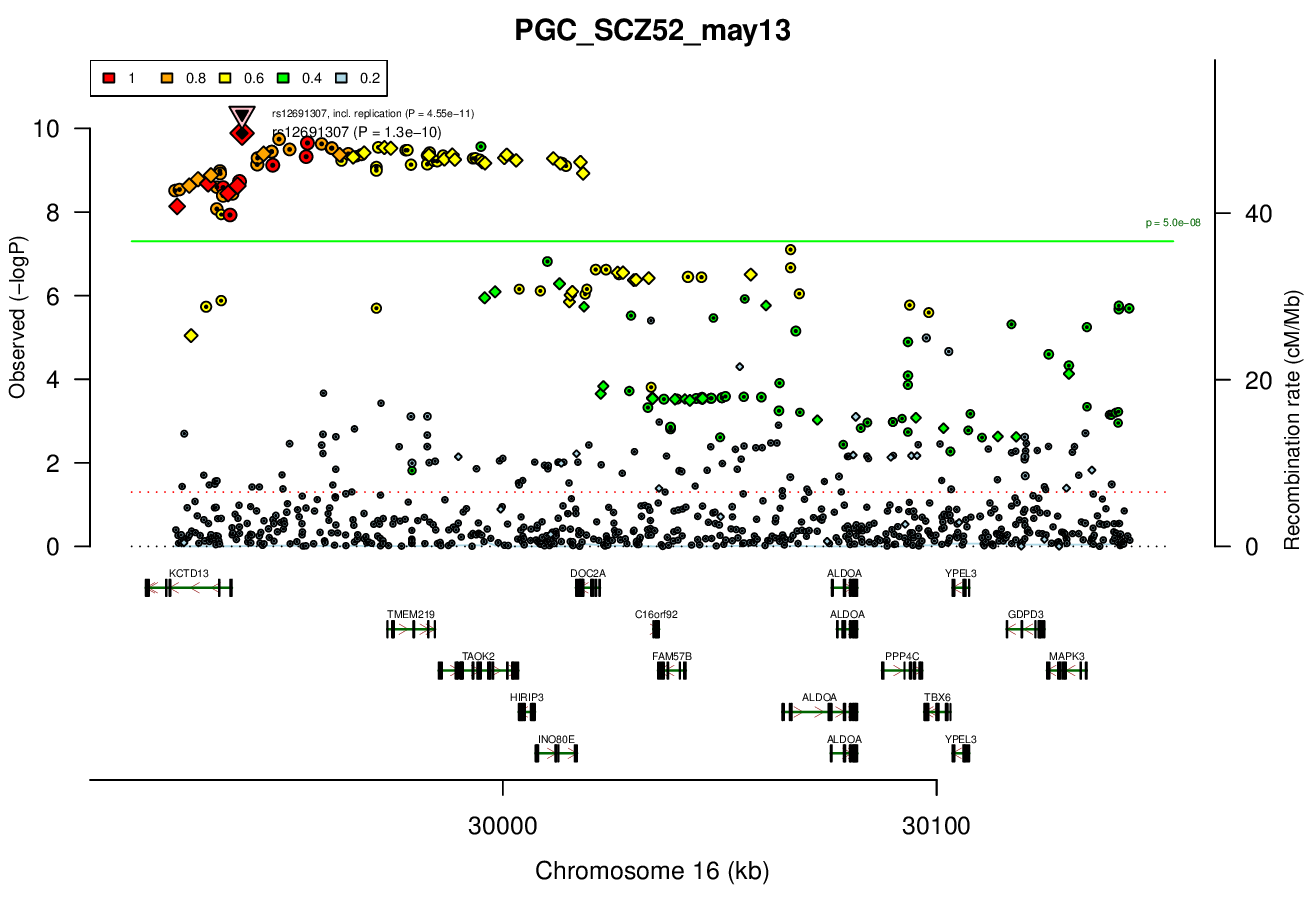
Gwas And Surrounding Region Pic
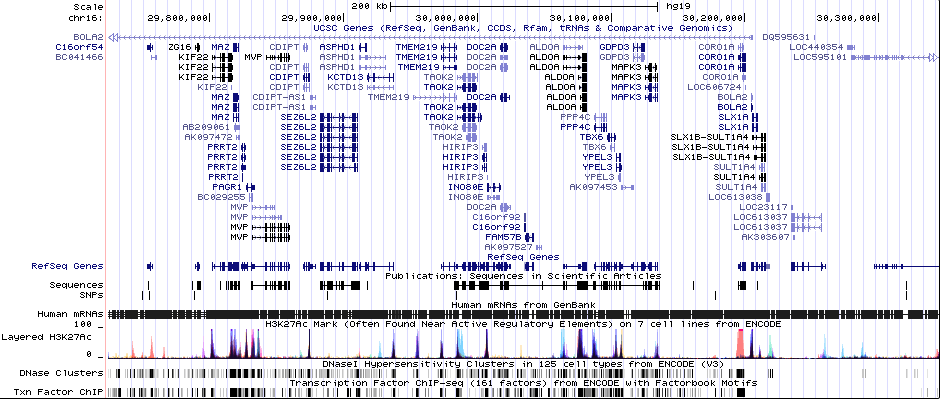
Gwas Region Pic
Sequence 1
Ensembl Gene Name
sez6l2Harvard Allele
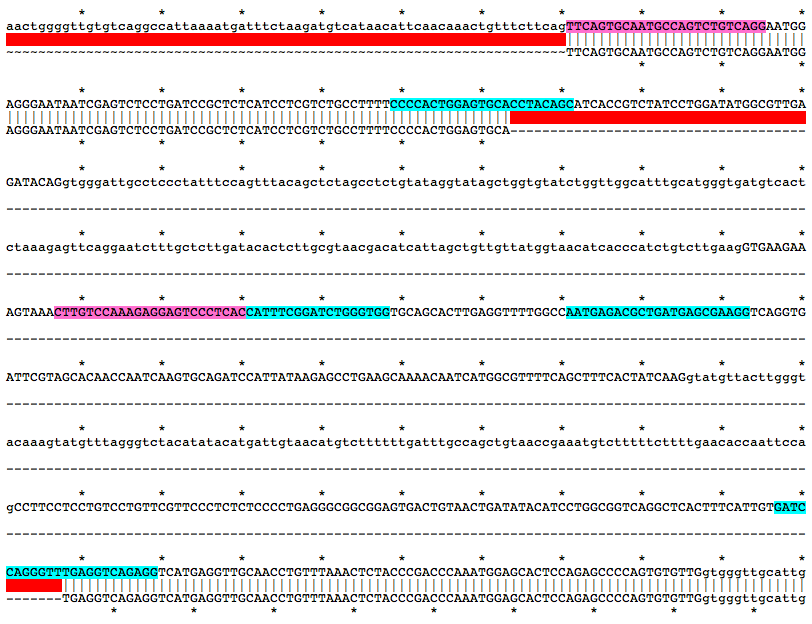
a328Mutation Area - WT DNA Sequence
TTCAGTGCAATGCCAGTCTGTCAGGAATGGAGGGAATAATCGAGTCTCCTGATCCGCTCTCATCCTCGTCTGCCTTTTCCCCACTGGAGTGCACCTACAGCATCACCGTCTATCCTGGATATGGCGTTGAGATACAGgtgggattgcctccctatttccagtttacagctctagcctctgtataggtatagctggtgtatctggttggcatttgcatgggtgatgtcactctaaagagttcaggaatctttgctcttgatacactcttgcgtaacgacatcattagctgttgttatggtaacatcacccatctgtcttgaagGTGAAGAAAGTAAACTTGTCCAAAGAGGAGTCCCTCACCATTTCGGATCTGGGTGGTGCAGCACTTGAGGTTTTGGCCAATGAGACGCTGATGAGCGAAGGTCAGGTGATTCGTAGCACAACCAATCAAGTGCAGATCCATTATAAGAGCCTGAAGCAAAACAATCATGGCGTTTTCAGCTTTCACTATCAAGgtatgttacttgggtacaaagtatgtttagggtctacatatacatgattgtaacatgtcttttttgatttgccagctgtaaccgaaatgtctttttcttttgaacaccaattccagCCTTCCTCCTGTCCTGTTCGTTCCCTCTCTCCCCTGAGGGCGGCGGAGTGACTGTAACTGATATACATCCTGGCGGTCAGGCTCACTTTCATTGTGATCCAGGGTTTGAGGTCAGAGGTCATGAGGTTGCAACCTGTTTAAACTCTACCCGACCCAAATGGAGCACTCCAGAGCCCCAGTGTGTTGgtgggttgcattgatttttgttatttgaatgtctttcatttgctcaaatacacctctgctcgttgccatgtgtacMutation Area - Mutant DNA Sequence
TTCAGTGCAATGCCAGTCTGTCAGGAATGGAGGGAATAATCGAGTCTCCTGATCCGCTCTCATCCTCGTCTGCCTTTTCCCCACTGGAGTGCATGAGGTCAGAGGTCATGAGGTTGCAACCTGTTTAAACTCTACCCGACCCAAATGGAGCACTCCAGAGCCCCAGTGTGTTGgtgggttgcattgatttttgttatttgaatgtctttcatttgctcaaatacacctctgctcgttgccatgtgtacGuide RNA Target Sites
GCTGTAGGTGCACTCCAGTGGGG
CTCACCATTTCGGATCTGGGTGG
AATGAGACGCTGATGAGCGAAGG
GATCCAGGGTTTGAGGTCAGAGG
Genotyping Primers
f, TTCAGTGCAATGCCAGTCTGTCAGG
r, gtacacatggcaacgagcagaggtg
wt_r, GTGAGGGACTCCTCTTTGGACAAG
Alignment of WT and Mutant Sequences w/ gRNA Targets (cyan) and Genotyping Primers (pink) Shown on WT Sequence
Allele
644bpDWT Genotyping Size
360WT Protein Sequence
MVSTVFAVTLSVTLLHLASGASLGTSDPEAPPTVTSSPRPLGDLIHAALLSKEYLGHTPG
SRGTTTNPTQAVPSIKESEPASTVMSPGILTTAVTSSAVPQAGQLGFGGAVTSLPAEEET
TTTLITTTTITTVHTPVQCNASLSGMEGIIESPDPLSSSSAFSPLECTYSITVYPGYGVE
IQVKKVNLSKEESLTISDLGGAALEVLANETLMSEGQVIRSTTNQVQIHYKSLKQNNHGV
FSFHYQAFLLSCSFPLSPEGGGVTVTDIHPGGQAHFHCDPGFEVRGHEVATCLNSTRPKW
STPEPQCVAVSCGGWIRNATVGRILSPTLPSASNHSSGSNLSCHWLLEAKEGHRLHLHFE
RVALDEDNDKLIVRSGNSTTAPLLFDSDLDDVPERGFVSEGSSLFVELTADSSSIPLLLA
LRYEAFDGEHCYEPYLPHGNFSSSDITFPLGTVVSFSCAAGFVMEQGSGVMECLDPNDPH
WNETEPVCRALCGGELTDPVGTVLSPDWPQSYSKGQDCVWQIHVSEDKRIELDVQILNIR
HNDVLTLFDGHDLMSHVIGQYMGSRERFRVVSGGSEVTIQFQSDPDDSTFILSQGFLIHY
REVEPNDTCSALPQIEFGWSSSSHPSLVRGSVLTYQCQPGYDIVGSDIITCQWDLTWSNS
PPTCVKIQQCPDPGEVVNGARSVHPESGFAVGTVVRFTCNQGYQLEGPSQISCHGRDTGM
PKWSDRSPKCVLKYDPCPNPGVPDNGYQTLYKHSYQAGETLRFFCYEGYELIGEVIINCV
PGHPSQWNSPPPFCKVAYEGLLDDHKLEVSQSLDASHQMLSENIALAIILPIILVILLIG
GIYMYYTNVCRWQWKPMFWKSLSHTHSYSPITVESDFNNPLYEAGDTREYEVSI-
Mutant Protein Sequence
MVSTVFAVTLSVTLLHLASGASLGTSDPEAPPTVTSSPRPLGDLIHAALLSKEYLGHTPG
SRGTTTNPTQAVPSIKESEPASTVMSPGILTTAVTSSAVPQAGQLGFGGAVTSLPAEEET
TTTLITTTTITTVHTPVQCNASLSGMEGIIESPDPLSSSSAFSPLECMRSEVMRLQPV-
Mutant Genotyping Size
248Ribosome Profiling Development
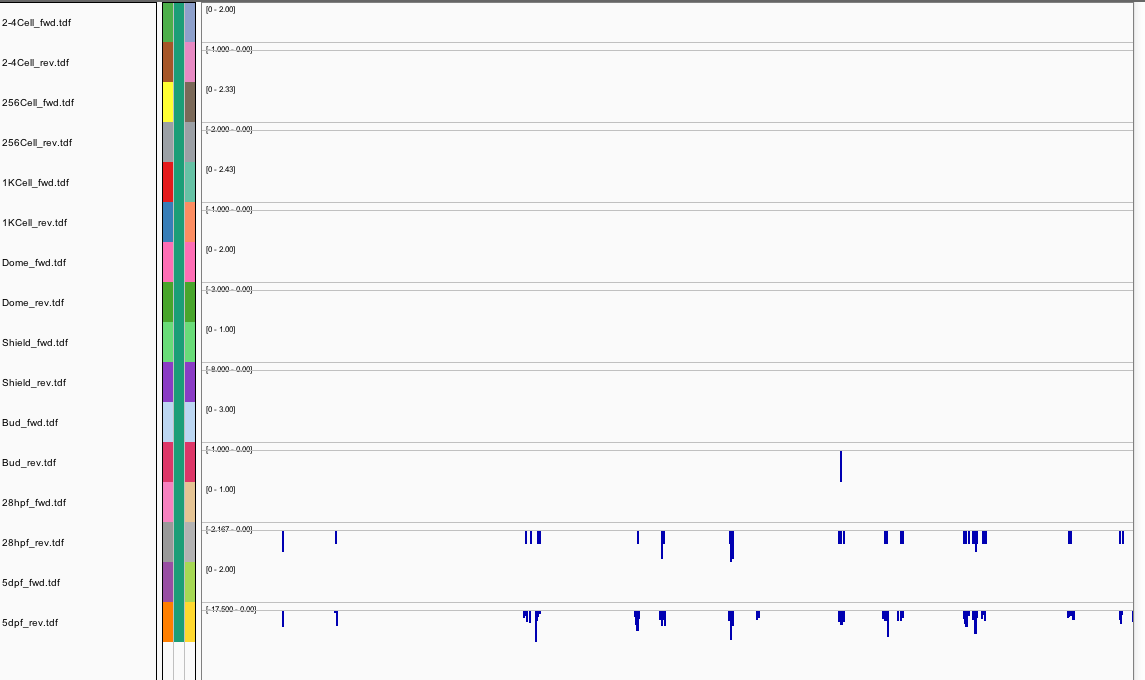
Behavior Data
Behavior Data Summary
NoneBehavior Data Description 1
Dark flash block 1 start Merged section Pvalue = not significant 17 het vs 25 hom ribgraph_mean_ribbon_fullboutdata_dpix_a0darkflash103.pngBehavior Data Graph 1
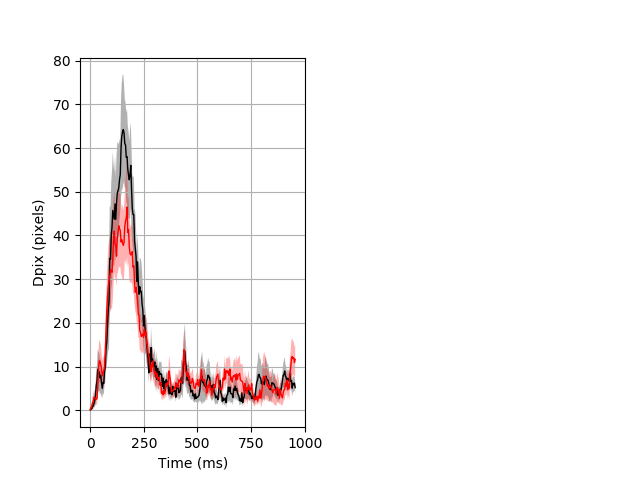
Behavior Data Description 2
Day all prepulse tap Merged section Pvalue = 0.0382 17 het vs 25 hom ribgraph_mean_ribbon_fullboutdata_dpix_dayprepulseinhibition100d.pngBehavior Data Graph 2
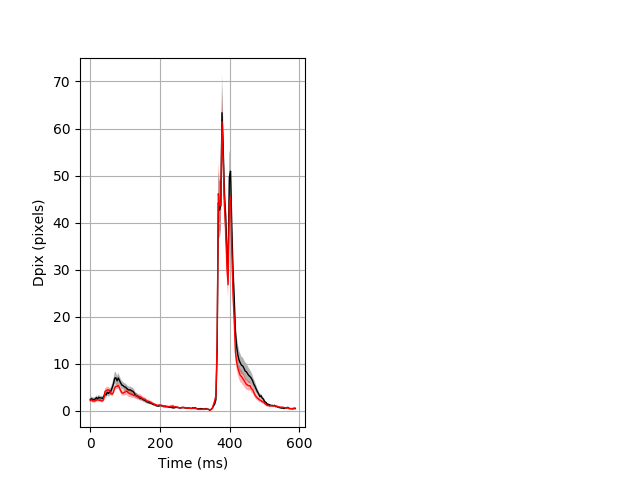
Behavior Data Description 3
Features of movement entire protocol Graph Pvalue = not significant Merged section Pvalue = not significant 17 het vs 25 hom ribgraph_mean_ribbonbout_aveboutdisp_10min_combo.pngBehavior Data Graph 3
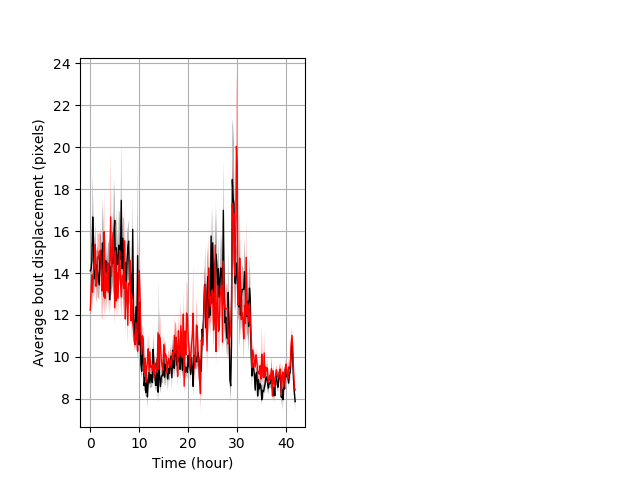
Behavior Data Description 4
Frequency of movement entire protocol Graph Pvalue = not significant Merged section Pvalue = not significant 17 het vs 25 hom ribgraph_mean_ribbonbout_dpixnumberofbouts_10min_combo.pngBehavior Data Graph 4
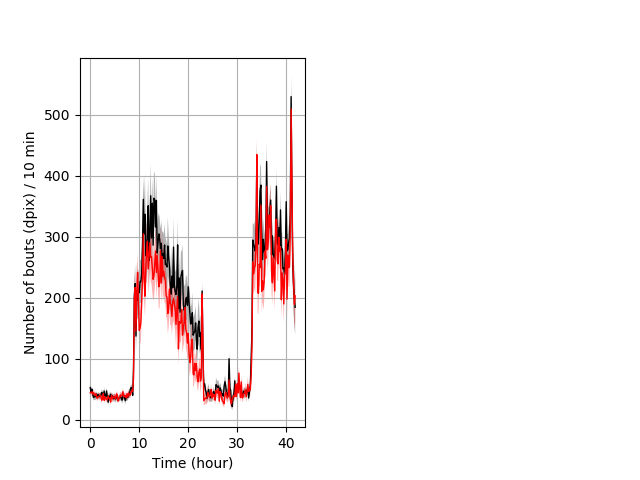
Behavior Data Description 5
Location in well entire protocol Graph Pvalue = not significant Merged section Pvalue = not significant 17 het vs 25 hom ribgraph_mean_ribbonbout_averhofrac_10min_combo.pngBehavior Data Graph 5
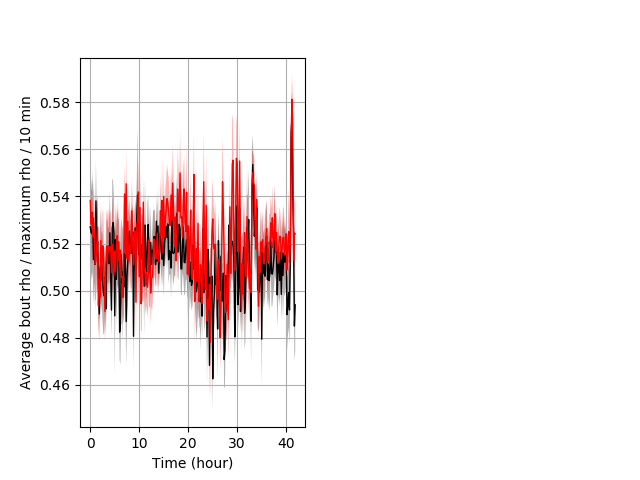
Behavior Data Description 6
NoneBehavior Data Graph 6
NoneBehavior Data Description 7
NoneBehavior Data Graph 7
NoneBehavior Data Description 8
NoneBehavior Data Graph 8
NoneBehavior Data Description 9
NoneBehavior Data Graph 9
NoneBehavior Data Description 10
NoneBehavior Data Graph 10
NoneBehavior Data Description 11
NoneBehavior Data Graph 11
NoneBehavior Data Description 12
NoneBehavior Data Graph 12
NoneBehavior Data Description 13
NoneBehavior Data Graph 13
NoneBehavior Data Description 14
NoneBehavior Data Graph 14
None